bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2919_orf2
Length=112
Score E
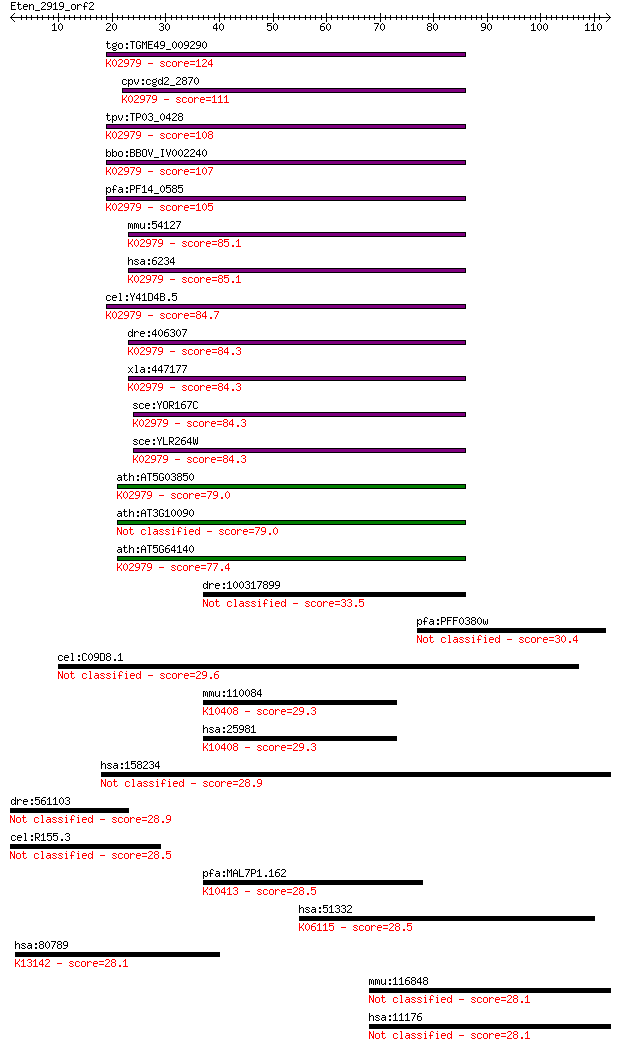
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_009290 40S ribosomal protein S28, putative ; K02979... 124 1e-28
cpv:cgd2_2870 40S ribosomal protein S28, no good start Met ; K... 111 7e-25
tpv:TP03_0428 40S ribosomal protein S28e; K02979 small subunit... 108 3e-24
bbo:BBOV_IV002240 21.m02745; 40S ribosomal protein S28; K02979... 107 8e-24
pfa:PF14_0585 40S ribosomal protein S28e, putative; K02979 sma... 105 4e-23
mmu:54127 Rps28; ribosomal protein S28; K02979 small subunit r... 85.1 5e-17
hsa:6234 RPS28; ribosomal protein S28; K02979 small subunit ri... 85.1 5e-17
cel:Y41D4B.5 rps-28; Ribosomal Protein, Small subunit family m... 84.7 6e-17
dre:406307 rps28, wu:fb07a10, wu:fb94f01, zgc:73367; ribosomal... 84.3 8e-17
xla:447177 rps28p9, MGC116486, MGC85550; ribosomal protein S28... 84.3 9e-17
sce:YOR167C RPS28A, RPS33A; Rps28ap; K02979 small subunit ribo... 84.3 9e-17
sce:YLR264W RPS28B, RPS33B; Rps28bp; K02979 small subunit ribo... 84.3 9e-17
ath:AT5G03850 40S ribosomal protein S28 (RPS28B); K02979 small... 79.0 4e-15
ath:AT3G10090 40S ribosomal protein S28 (RPS28A) 79.0 4e-15
ath:AT5G64140 RPS28; RPS28 (RIBOSOMAL PROTEIN S28); structural... 77.4 1e-14
dre:100317899 dnah1; dynein, axonemal, heavy chain 1 33.5
pfa:PFF0380w conserved Plasmodium protein, unknown function 30.4 1.6
cel:C09D8.1 ptp-3; Protein Tyrosine Phosphatase family member ... 29.6 2.3
mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034... 29.3 3.4
hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,... 29.3 3.5
hsa:158234 RG9MTD3, FLJ31455, RP11-3J10.9, bA3J10.9; RNA (guan... 28.9 3.7
dre:561103 hypothetical LOC561103 28.9 4.5
cel:R155.3 hypothetical protein 28.5 4.8
pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein hea... 28.5 5.1
hsa:51332 SPTBN5, BSPECV, HUBSPECV, HUSPECV; spectrin, beta, n... 28.5 5.6
hsa:80789 INTS5, INT5, KIAA1698; integrator complex subunit 5;... 28.1 7.0
mmu:116848 Baz2a, AA415431, C030005G16Rik, C78388, Tip5, Walp3... 28.1 7.5
hsa:11176 BAZ2A, DKFZp781B109, FLJ13768, FLJ13780, FLJ45876, K... 28.1 7.8
> tgo:TGME49_009290 40S ribosomal protein S28, putative ; K02979
small subunit ribosomal protein S28e
Length=68
Score = 124 bits (310), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 62/67 (92%), Positives = 64/67 (95%), Gaps = 0/67 (0%)
Query 19 MEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLET 78
MEQPKLAKVEKVLGRTGSRGGV QVRVTFM +E+DL GRSLIRNVKGPVREGDILALLET
Sbjct 1 MEQPKLAKVEKVLGRTGSRGGVLQVRVTFMADETDLAGRSLIRNVKGPVREGDILALLET 60
Query 79 EREARRL 85
EREARRL
Sbjct 61 EREARRL 67
> cpv:cgd2_2870 40S ribosomal protein S28, no good start Met ;
K02979 small subunit ribosomal protein S28e
Length=68
Score = 111 bits (277), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 56/64 (87%), Positives = 60/64 (93%), Gaps = 0/64 (0%)
Query 22 PKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETERE 81
PKLAKVEK+LGRTGSRGGV QVRV FM+ ES+L GRSLIRNV+GPVREGDILALLETERE
Sbjct 4 PKLAKVEKILGRTGSRGGVIQVRVQFMSGESELAGRSLIRNVRGPVREGDILALLETERE 63
Query 82 ARRL 85
ARRL
Sbjct 64 ARRL 67
> tpv:TP03_0428 40S ribosomal protein S28e; K02979 small subunit
ribosomal protein S28e
Length=67
Score = 108 bits (271), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 57/67 (85%), Positives = 60/67 (89%), Gaps = 1/67 (1%)
Query 19 MEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLET 78
MEQPKLAKV KVLGRTG+RGGV QVRV FM E S +GR+LIRNVKGPVREGDILALLET
Sbjct 1 MEQPKLAKVNKVLGRTGNRGGVTQVRVDFMGE-SGWEGRTLIRNVKGPVREGDILALLET 59
Query 79 EREARRL 85
EREARRL
Sbjct 60 EREARRL 66
> bbo:BBOV_IV002240 21.m02745; 40S ribosomal protein S28; K02979
small subunit ribosomal protein S28e
Length=67
Score = 107 bits (267), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 57/67 (85%), Positives = 58/67 (86%), Gaps = 1/67 (1%)
Query 19 MEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLET 78
MEQPKLAKV KVLGRTGSRGGV QVRV FM E +GR LIRNVKGPVREGDILALLET
Sbjct 1 MEQPKLAKVNKVLGRTGSRGGVTQVRVDFMGE-GGWEGRVLIRNVKGPVREGDILALLET 59
Query 79 EREARRL 85
EREARRL
Sbjct 60 EREARRL 66
> pfa:PF14_0585 40S ribosomal protein S28e, putative; K02979 small
subunit ribosomal protein S28e
Length=67
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 56/67 (83%), Positives = 59/67 (88%), Gaps = 1/67 (1%)
Query 19 MEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLET 78
ME+ KLAKVEKVLGRTGSRGGV QVR FM + S+L GR LIRNVKGPVREGDILALLET
Sbjct 1 MEKSKLAKVEKVLGRTGSRGGVIQVRAQFMGD-SELAGRFLIRNVKGPVREGDILALLET 59
Query 79 EREARRL 85
EREARRL
Sbjct 60 EREARRL 66
> mmu:54127 Rps28; ribosomal protein S28; K02979 small subunit
ribosomal protein S28e
Length=69
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 45/63 (71%), Positives = 51/63 (80%), Gaps = 4/63 (6%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREA 82
KLA+V KVLGRTGS+G QVRV FM + S RS+IRNVKGPVREGD+L LLE+EREA
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVEFMDDTS----RSIIRNVKGPVREGDVLTLLESEREA 65
Query 83 RRL 85
RRL
Sbjct 66 RRL 68
> hsa:6234 RPS28; ribosomal protein S28; K02979 small subunit
ribosomal protein S28e
Length=69
Score = 85.1 bits (209), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 45/63 (71%), Positives = 51/63 (80%), Gaps = 4/63 (6%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREA 82
KLA+V KVLGRTGS+G QVRV FM + S RS+IRNVKGPVREGD+L LLE+EREA
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVEFMDDTS----RSIIRNVKGPVREGDVLTLLESEREA 65
Query 83 RRL 85
RRL
Sbjct 66 RRL 68
> cel:Y41D4B.5 rps-28; Ribosomal Protein, Small subunit family
member (rps-28); K02979 small subunit ribosomal protein S28e
Length=65
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 43/67 (64%), Positives = 55/67 (82%), Gaps = 3/67 (4%)
Query 19 MEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLET 78
M++ LA+V KV+GRTGS+G QVRV F+ ++++ RS+IRNVKGPVREGDIL LLE+
Sbjct 1 MDKLTLARVTKVIGRTGSQGQCTQVRVEFINDQNN---RSIIRNVKGPVREGDILTLLES 57
Query 79 EREARRL 85
EREARRL
Sbjct 58 EREARRL 64
> dre:406307 rps28, wu:fb07a10, wu:fb94f01, zgc:73367; ribosomal
protein S28; K02979 small subunit ribosomal protein S28e
Length=69
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 45/63 (71%), Positives = 50/63 (79%), Gaps = 4/63 (6%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREA 82
KLA+V KVLGRTGS+G QVRV FM D RS+IRNVKGPVREGD+L LLE+EREA
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVEFM----DDSNRSIIRNVKGPVREGDVLTLLESEREA 65
Query 83 RRL 85
RRL
Sbjct 66 RRL 68
> xla:447177 rps28p9, MGC116486, MGC85550; ribosomal protein S28
pseudogene 9; K02979 small subunit ribosomal protein S28e
Length=69
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 45/63 (71%), Positives = 50/63 (79%), Gaps = 4/63 (6%)
Query 23 KLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREA 82
KLA+V KVLGRTGS+G QVRV FM D RS+IRNVKGPVREGD+L LLE+EREA
Sbjct 10 KLARVTKVLGRTGSQGQCTQVRVEFM----DDSNRSIIRNVKGPVREGDVLTLLESEREA 65
Query 83 RRL 85
RRL
Sbjct 66 RRL 68
> sce:YOR167C RPS28A, RPS33A; Rps28ap; K02979 small subunit ribosomal
protein S28e
Length=67
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 44/62 (70%), Positives = 51/62 (82%), Gaps = 4/62 (6%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREAR 83
LAKV KVLGRTGSRGGV QVRV F+ + S R+++RNVKGPVRE DIL L+E+EREAR
Sbjct 9 LAKVIKVLGRTGSRGGVTQVRVEFLEDTS----RTIVRNVKGPVRENDILVLMESEREAR 64
Query 84 RL 85
RL
Sbjct 65 RL 66
> sce:YLR264W RPS28B, RPS33B; Rps28bp; K02979 small subunit ribosomal
protein S28e
Length=67
Score = 84.3 bits (207), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 44/62 (70%), Positives = 51/62 (82%), Gaps = 4/62 (6%)
Query 24 LAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREAR 83
LAKV KVLGRTGSRGGV QVRV F+ + S R+++RNVKGPVRE DIL L+E+EREAR
Sbjct 9 LAKVIKVLGRTGSRGGVTQVRVEFLEDTS----RTIVRNVKGPVRENDILVLMESEREAR 64
Query 84 RL 85
RL
Sbjct 65 RL 66
> ath:AT5G03850 40S ribosomal protein S28 (RPS28B); K02979 small
subunit ribosomal protein S28e
Length=64
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 45/65 (69%), Positives = 50/65 (76%), Gaps = 5/65 (7%)
Query 21 QPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETER 80
Q K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGDIL LLE+ER
Sbjct 4 QIKHAVVVKVMGRTGSRGQVTQVRVKF--TDSD---RYIMRNVKGPVREGDILTLLESER 58
Query 81 EARRL 85
EARRL
Sbjct 59 EARRL 63
> ath:AT3G10090 40S ribosomal protein S28 (RPS28A)
Length=64
Score = 79.0 bits (193), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 45/65 (69%), Positives = 50/65 (76%), Gaps = 5/65 (7%)
Query 21 QPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETER 80
Q K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGDIL LLE+ER
Sbjct 4 QIKHAVVVKVMGRTGSRGQVTQVRVKF--TDSD---RYIMRNVKGPVREGDILTLLESER 58
Query 81 EARRL 85
EARRL
Sbjct 59 EARRL 63
> ath:AT5G64140 RPS28; RPS28 (RIBOSOMAL PROTEIN S28); structural
constituent of ribosome; K02979 small subunit ribosomal protein
S28e
Length=64
Score = 77.4 bits (189), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 44/65 (67%), Positives = 50/65 (76%), Gaps = 5/65 (7%)
Query 21 QPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETER 80
Q K A V KV+GRTGSRG V QVRV F +SD R ++RNVKGPVREGD+L LLE+ER
Sbjct 4 QIKHAVVVKVMGRTGSRGQVTQVRVKF--TDSD---RFIMRNVKGPVREGDVLTLLESER 58
Query 81 EARRL 85
EARRL
Sbjct 59 EARRL 63
> dre:100317899 dnah1; dynein, axonemal, heavy chain 1
Length=1874
Score = 33.5 bits (75), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 37 RGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLETEREARRL 85
+ G+Q V++TF+ ++ ++ S + +V + GD+ L +E + R L
Sbjct 234 KAGMQNVQITFLFVDTQIKSESFLEDVNNILNSGDVPNLYTSEEQERIL 282
> pfa:PFF0380w conserved Plasmodium protein, unknown function
Length=2752
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 17/35 (48%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 77 ETEREARRLQRRQRLQRRQLLQRRQLLQRRQLLQR 111
+T LQR Q LQR LQR LQR LQR
Sbjct 1057 DTPTRGENLQRGQNLQRGDNLQRGDNLQRGDNLQR 1091
> cel:C09D8.1 ptp-3; Protein Tyrosine Phosphatase family member
(ptp-3)
Length=2227
Score = 29.6 bits (65), Expect = 2.3, Method: Composition-based stats.
Identities = 24/97 (24%), Positives = 41/97 (42%), Gaps = 0/97 (0%)
Query 10 PQTLAPNPKMEQPKLAKVEKVLGRTGSRGGVQQVRVTFMTEESDLQGRSLIRNVKGPVRE 69
PQ + P V K++ +G++ G Q +R+T TEE I V
Sbjct 1301 PQEVVTITDPTGPPFVDVPKLVDSSGTQPGQQMIRLTPATEEYGPISHYWIILVPANYST 1360
Query 70 GDILALLETEREARRLQRRQRLQRRQLLQRRQLLQRR 106
D++ L E E ++R +L R + + L+R+
Sbjct 1361 EDVVNLDPIELEKATAEKRAQLARSLSVSPSKKLKRK 1397
> mmu:110084 Dnahc1, B230373P09Rik, DKFZp434A236, Dnah1, E030034C22Rik,
MDHC7, MGC37121; dynein, axonemal, heavy chain 1;
K10408 dynein heavy chain, axonemal
Length=4250
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 8/36 (22%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 37 RGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDI 72
+ G+Q + +TF+ ++ ++ S + ++ + GDI
Sbjct 2616 KAGLQNLPITFLFSDTQIKNESFLEDINNVLNSGDI 2651
> hsa:25981 DNAH1, DNAHC1, HDHC7, HL-11, HL11, XLHSRF-1; dynein,
axonemal, heavy chain 1; K10408 dynein heavy chain, axonemal
Length=4265
Score = 29.3 bits (64), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 8/36 (22%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 37 RGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDI 72
+ G+Q + +TF+ ++ ++ S + ++ + GDI
Sbjct 2631 KAGLQNLPITFLFSDTQIKNESFLEDINNVLNSGDI 2666
> hsa:158234 RG9MTD3, FLJ31455, RP11-3J10.9, bA3J10.9; RNA (guanine-9-)
methyltransferase domain containing 3
Length=316
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 46/98 (46%), Gaps = 21/98 (21%)
Query 18 KMEQPKLAKVEKVLGRTGSRG---GVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILA 74
K+E P L E +L TG G G Q +++ + +G +EG+ILA
Sbjct 11 KVESPVLQGQEGILEETGEDGLPEGFQLLQI----------------DAEGECQEGEILA 54
Query 75 LLETEREARRLQRRQRLQRRQLLQRRQLLQRRQLLQRR 112
T ++ +QR+QR + + ++ +R+Q +RR
Sbjct 55 TGSTAWCSKNVQRKQRHWEKIVAAKKS--KRKQEKERR 90
> dre:561103 hypothetical LOC561103
Length=344
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 10/22 (45%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 1 PQTPNPSPKPQTLAPNPKMEQP 22
PQ PS +P T PNP++++P
Sbjct 11 PQPTQPSYQPSTPKPNPELDKP 32
> cel:R155.3 hypothetical protein
Length=1165
Score = 28.5 bits (62), Expect = 4.8, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 15/28 (53%), Gaps = 0/28 (0%)
Query 1 PQTPNPSPKPQTLAPNPKMEQPKLAKVE 28
P P P P P P ++EQ +AKV+
Sbjct 1123 PIAPEPKPTPAETKPKDRLEQRGMAKVD 1150
> pfa:MAL7P1.162 dynein heavy chain, putative; K10413 dynein heavy
chain 1, cytosolic
Length=4985
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 10/41 (24%), Positives = 25/41 (60%), Gaps = 0/41 (0%)
Query 37 RGGVQQVRVTFMTEESDLQGRSLIRNVKGPVREGDILALLE 77
R G+++ ++TF+ +ES++ G + + + + G++ L E
Sbjct 3180 RAGIKEEKITFIFDESNVLGPAFLERMNALLASGEVPGLFE 3220
> hsa:51332 SPTBN5, BSPECV, HUBSPECV, HUSPECV; spectrin, beta,
non-erythrocytic 5; K06115 spectrin beta
Length=3639
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 1/55 (1%)
Query 55 QGRSLIRNVKGPVREGDILALLETEREARRLQRRQRLQRRQLLQRRQLLQRRQLL 109
Q +L+ + VREG LA + E +ARRL +R + R L +RR L+ R LL
Sbjct 2807 QAEALLGQAQAFVREGHCLAQ-DVEEQARRLLQRFKSLREPLQERRTALEARSLL 2860
> hsa:80789 INTS5, INT5, KIAA1698; integrator complex subunit
5; K13142 integrator complex subunit 5
Length=1019
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 2 QTPNPSPKPQTLAPNPKMEQPKLAKVEKVLGRTGSRGG 39
QTP+ P P + A + PK+A V +LG SR G
Sbjct 269 QTPSTDPFPGSPAIPAEKRVPKIASVVGILGHLASRHG 306
> mmu:116848 Baz2a, AA415431, C030005G16Rik, C78388, Tip5, Walp3,
mKIAA0314; bromodomain adjacent to zinc finger domain, 2A
Length=1887
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 4/49 (8%)
Query 68 REGDI----LALLETEREARRLQRRQRLQRRQLLQRRQLLQRRQLLQRR 112
+EG+I ++ E ER +L +RQ R++LL Q+L+ L Q R
Sbjct 1061 KEGEIDVAASSIPELERHIEKLSKRQLFFRKKLLHSSQMLRAVSLGQDR 1109
> hsa:11176 BAZ2A, DKFZp781B109, FLJ13768, FLJ13780, FLJ45876,
KIAA0314, TIP5, WALp3; bromodomain adjacent to zinc finger
domain, 2A
Length=1905
Score = 28.1 bits (61), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 68 REGDILA----LLETEREARRLQRRQRLQRRQLLQRRQLLQRRQLLQRR 112
R+G++ A + E ER+ +L +RQ R++LL Q+L+ L Q R
Sbjct 1072 RDGEVDATASSIPELERQIEKLSKRQLFFRKKLLHSSQMLRAVSLGQDR 1120
Lambda K H
0.318 0.134 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2057481480
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40