bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2879_orf1
Length=152
Score E
Sequences producing significant alignments: (Bits) Value
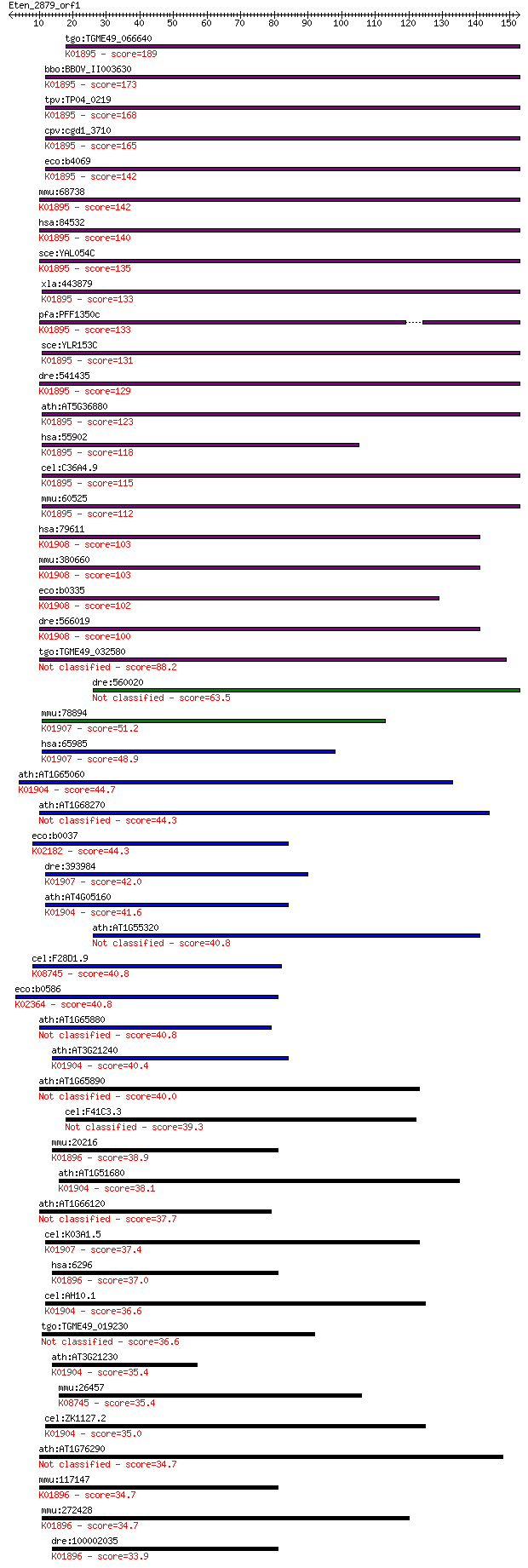
tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6... 189 4e-48
bbo:BBOV_II003630 18.m06304; acetyl-CoA synthetase (EC:6.2.1.1... 173 2e-43
tpv:TP04_0219 acetyl-coenzyme A synthetase (EC:6.2.1.1); K0189... 168 5e-42
cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA... 165 6e-41
eco:b4069 acs, acsA, ECK4062, JW4030, yfaC; acetyl-CoA synthet... 142 4e-34
mmu:68738 Acss1, 1110032O15Rik, AI788978, Acas2, Acas2l, AceCS... 142 5e-34
hsa:84532 ACSS1, ACAS2L, AceCS2L, FLJ45659, MGC33843; acyl-CoA... 140 2e-33
sce:YAL054C ACS1, FUN44; Acetyl-coA synthetase isoform which, ... 135 4e-32
xla:443879 acss2.2, MGC80104, acas2, acecs, acs, acsa, acss2; ... 133 2e-31
pfa:PFF1350c acetyl-CoA synthetase (EC:6.2.1.1); K01895 acetyl... 133 2e-31
sce:YLR153C ACS2; Acetyl-coA synthetase isoform which, along w... 131 1e-30
dre:541435 acss1, MGC158738, MGC174587, cb395, fi28d10, sb:cb3... 129 4e-30
ath:AT5G36880 acetyl-CoA synthetase, putative / acetate-CoA li... 123 2e-28
hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161... 118 8e-27
cel:C36A4.9 hypothetical protein; K01895 acetyl-CoA synthetase... 115 7e-26
mmu:60525 Acss2, 1110017C11Rik, ACAS, Acas1, Acas2, AceCS1, Ac... 112 6e-25
hsa:79611 ACSS3, FLJ21963; acyl-CoA synthetase short-chain fam... 103 2e-22
mmu:380660 Acss3, 8430416H19Rik, Gm874; acyl-CoA synthetase sh... 103 2e-22
eco:b0335 prpE, ECK0332, JW0326, yahU; propionate--CoA ligase;... 102 6e-22
dre:566019 acss3, si:dkey-231a18.1; acyl-CoA synthetase short-... 100 2e-21
tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6... 88.2 1e-17
dre:560020 hypothetical LOC560020 63.5 2e-10
mmu:78894 Aacs, 2210408B16Rik, SUR5; acetoacetyl-CoA synthetas... 51.2 1e-06
hsa:65985 AACS, ACSF1, FLJ12389, FLJ41251, SUR-5; acetoacetyl-... 48.9 7e-06
ath:AT1G65060 4CL3; 4CL3; 4-coumarate-CoA ligase (EC:6.2.1.12)... 44.7 1e-04
ath:AT1G68270 AMP-dependent synthetase and ligase family protein 44.3 1e-04
eco:b0037 caiC, ECK0038, JW0036, yaaM; probable crotonobetaine... 44.3 2e-04
dre:393984 aacs, MGC56105, zgc:56105; acetoacetyl-CoA syntheta... 42.0 7e-04
ath:AT4G05160 4-coumarate--CoA ligase, putative / 4-coumaroyl-... 41.6 0.001
ath:AT1G55320 AAE18; AAE18 (ACYL-ACTIVATING ENZYME 18); cataly... 40.8 0.002
cel:F28D1.9 hypothetical protein; K08745 solute carrier family... 40.8 0.002
eco:b0586 entF, ECK0579, JW0578; enterobactin synthase multien... 40.8 0.002
ath:AT1G65880 BZO1; BZO1 (BENZOYLOXYGLUCOSINOLATE 1); benzoate... 40.8 0.002
ath:AT3G21240 4CL2; 4CL2 (4-COUMARATE:COA LIGASE 2); 4-coumara... 40.4 0.002
ath:AT1G65890 AAE12; AAE12 (ACYL ACTIVATING ENZYME 12); catalytic 40.0 0.003
cel:F41C3.3 acs-11; fatty Acid CoA Synthetase family member (a... 39.3 0.005
mmu:20216 Acsm3, Sa, Sah; acyl-CoA synthetase medium-chain fam... 38.9 0.007
ath:AT1G51680 4CL1; 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumara... 38.1 0.011
ath:AT1G66120 acyl-activating enzyme 11 (AAE11) 37.7 0.013
cel:K03A1.5 sur-5; SUppressor of activated let-60 Ras family m... 37.4 0.020
hsa:6296 ACSM3, SA, SAH; acyl-CoA synthetase medium-chain fami... 37.0 0.023
cel:AH10.1 hypothetical protein; K01904 4-coumarate--CoA ligas... 36.6 0.032
tgo:TGME49_019230 acetoacetyl-CoA synthetase, putative (EC:6.2... 36.6 0.034
ath:AT3G21230 4CL5; 4CL5 (4-coumarate:CoA ligase 5); 4-coumara... 35.4 0.067
mmu:26457 Slc27a1, FATP1, Fatp; solute carrier family 27 (fatt... 35.4 0.068
cel:ZK1127.2 hypothetical protein; K01904 4-coumarate--CoA lig... 35.0 0.086
ath:AT1G76290 AMP-dependent synthetase and ligase family protein 34.7 0.11
mmu:117147 Acsm1, Acas3, Bucs1, Macs; acyl-CoA synthetase medi... 34.7 0.12
mmu:272428 Acsm5, C730019D22, C730027J19Rik; acyl-CoA syntheta... 34.7 0.12
dre:100002035 acsm3, zgc:172040; acyl-CoA synthetase medium-ch... 33.9 0.23
> tgo:TGME49_066640 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=535
Score = 189 bits (479), Expect = 4e-48, Method: Composition-based stats.
Identities = 86/136 (63%), Positives = 111/136 (81%), Gaps = 1/136 (0%)
Query 18 LLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHSRV 77
+LK L VRKGD+V+IYLPM PEIVYAMLACARIGAVH+V+FAGF+A+SLAER+ DA S +
Sbjct 1 MLKRLGVRKGDVVSIYLPMGPEIVYAMLACARIGAVHNVIFAGFAASSLAERIHDAESHI 60
Query 78 LITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEP-SSFVEGRDLSGDDLLPQ 136
LITA + RGG+ + LK + DEA+ +C V+H+V+FR EP ++VEGRD+ G++L+ Q
Sbjct 61 LITADQSSRGGKTIFLKDIADEAMAICPQVRHLVVFRCKGEPLKNWVEGRDIWGNELMSQ 120
Query 137 MRPYCPLETMDSEDML 152
M+PYCPLE MDSED L
Sbjct 121 MQPYCPLEVMDSEDPL 136
> bbo:BBOV_II003630 18.m06304; acetyl-CoA synthetase (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=703
Score = 173 bits (439), Expect = 2e-43, Method: Composition-based stats.
Identities = 78/141 (55%), Positives = 103/141 (73%), Gaps = 0/141 (0%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
VCRFAN+L VRKGD+VT+Y+P +PE+ YAMLACARIGA+HSVVF G+SA S+AER+
Sbjct 162 VCRFANVLIDKGVRKGDVVTLYMPSIPELAYAMLACARIGAIHSVVFGGYSAASIAERIR 221
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLSGD 131
DA S ++IT + RGG+ +++K +VDEAL C+ V ++ R + + EGRD D
Sbjct 222 DADSHIIITVDESYRGGKTIKMKSIVDEALLSCAGVTTCLVLRYAGVKVNMKEGRDFWLD 281
Query 132 DLLPQMRPYCPLETMDSEDML 152
DLL +RPYCP+E MDSED L
Sbjct 282 DLLEHVRPYCPIEVMDSEDSL 302
> tpv:TP04_0219 acetyl-coenzyme A synthetase (EC:6.2.1.1); K01895
acetyl-CoA synthetase [EC:6.2.1.1]
Length=681
Score = 168 bits (426), Expect = 5e-42, Method: Composition-based stats.
Identities = 76/141 (53%), Positives = 102/141 (72%), Gaps = 0/141 (0%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
VC+ AN+LK L RKGD VTIY+P +PE+ Y++LACARIGAVHSVVF GFSA SLAER+
Sbjct 147 VCKVANVLKMLGARKGDCVTIYMPTIPELCYSVLACARIGAVHSVVFGGFSATSLAERIH 206
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLSGD 131
D++S ++ITA GMR + ++ K ++D+AL CS V H ++FR GRD+
Sbjct 207 DSNSHIVITADGGMRATKHIKTKDIMDDALTRCSFVTHCLVFRNIGSDVKMTPGRDVWMH 266
Query 132 DLLPQMRPYCPLETMDSEDML 152
+ + +RPYCP+ETMDSED+L
Sbjct 267 EAMESVRPYCPVETMDSEDVL 287
> cpv:cgd1_3710 acetyl-coenzyme A synthetase ; K01895 acetyl-CoA
synthetase [EC:6.2.1.1]
Length=695
Score = 165 bits (417), Expect = 6e-41, Method: Composition-based stats.
Identities = 74/141 (52%), Positives = 102/141 (72%), Gaps = 0/141 (0%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
VC+ AN+LK ++KGD V IY+PM+PE +Y MLACARIGAVH VVFAGF+A +L ERL
Sbjct 155 VCKMANVLKRFGIKKGDSVGIYMPMIPETIYTMLACARIGAVHMVVFAGFAAQNLLERLV 214
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLSGD 131
+A ++++TA +G RG +++ LK VVD+AL+ ++ ++FR P FV+GRD G+
Sbjct 215 NARCKIVVTADQGSRGKKIINLKDVVDKALEKIPEIKTCIVFRHLNGPIEFVKGRDFDGE 274
Query 132 DLLPQMRPYCPLETMDSEDML 152
L+ +PYCPLE MDSED L
Sbjct 275 TLMRSEKPYCPLEDMDSEDPL 295
> eco:b4069 acs, acsA, ECK4062, JW4030, yfaC; acetyl-CoA synthetase
(EC:6.2.1.1); K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=652
Score = 142 bits (359), Expect = 4e-34, Method: Composition-based stats.
Identities = 71/143 (49%), Positives = 100/143 (69%), Gaps = 2/143 (1%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
VCRFAN L L ++KGD+V IY+PM+PE AMLACARIGAVHSV+F GFS ++A R+
Sbjct 117 VCRFANTLLELGIKKGDVVAIYMPMVPEAAVAMLACARIGAVHSVIFGGFSPEAVAGRII 176
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALK--LCSCVQHVVLFRKSAEPSSFVEGRDLS 129
D++SR++IT+ +G+R GR + LK+ VD+ALK + V+HVV+ +++ + EGRDL
Sbjct 177 DSNSRLVITSDEGVRAGRSIPLKKNVDDALKNPNVTSVEHVVVLKRTGGKIDWQEGRDLW 236
Query 130 GDDLLPQMRPYCPLETMDSEDML 152
DL+ Q E M++ED L
Sbjct 237 WHDLVEQASDQHQAEEMNAEDPL 259
> mmu:68738 Acss1, 1110032O15Rik, AI788978, Acas2, Acas2l, AceCS2;
acyl-CoA synthetase short-chain family member 1 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=682
Score = 142 bits (357), Expect = 5e-34, Method: Composition-based stats.
Identities = 71/143 (49%), Positives = 96/143 (67%), Gaps = 1/143 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+ CR AN LK V +GD V IY+P+ P V AMLACARIGA+H+VVFAGFSA SLA R
Sbjct 141 ETTCRLANTLKRHGVHRGDRVAIYMPVSPLAVAAMLACARIGAIHTVVFAGFSAESLAGR 200
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLS 129
+ DA + +IT +G+RGGRVV LK++VDEA+K C VQHV++ ++ + + D+
Sbjct 201 INDAKCKAVITFNQGLRGGRVVELKKIVDEAVKSCPTVQHVLVAHRT-DTKVPMGSLDIP 259
Query 130 GDDLLPQMRPYCPLETMDSEDML 152
+ + + P C E+M SEDML
Sbjct 260 LEQEMAKEAPVCTPESMSSEDML 282
> hsa:84532 ACSS1, ACAS2L, AceCS2L, FLJ45659, MGC33843; acyl-CoA
synthetase short-chain family member 1 (EC:6.2.1.1); K01895
acetyl-CoA synthetase [EC:6.2.1.1]
Length=689
Score = 140 bits (352), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 72/143 (50%), Positives = 97/143 (67%), Gaps = 1/143 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+ CR AN LK V +GD V IY+P+ P V AMLACARIGAVH+V+FAGFSA SLA R
Sbjct 148 ETTCRLANTLKRHGVHRGDRVAIYMPVSPLAVAAMLACARIGAVHTVIFAGFSAESLAGR 207
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLS 129
+ DA +V+IT +G+RGGRVV LK++VDEA+K C VQHV++ ++ + + D+
Sbjct 208 INDAKCKVVITFNQGLRGGRVVELKKIVDEAVKHCPTVQHVLVAHRT-DNKVHMGDLDVP 266
Query 130 GDDLLPQMRPYCPLETMDSEDML 152
+ + + P C E+M SEDML
Sbjct 267 LEQEMAKEDPVCAPESMGSEDML 289
> sce:YAL054C ACS1, FUN44; Acetyl-coA synthetase isoform which,
along with Acs2p, is the nuclear source of acetyl-coA for
histone acetlyation; expressed during growth on nonfermentable
carbon sources and under aerobic conditions (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=713
Score = 135 bits (341), Expect = 4e-32, Method: Composition-based stats.
Identities = 67/145 (46%), Positives = 100/145 (68%), Gaps = 2/145 (1%)
Query 10 DQVCRFANLLK-SLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAE 68
++VC+ A +L S+ VRKGD V +Y+PM+PE + +LA +RIGA+HSVVFAGFS+NSL +
Sbjct 171 EEVCQVAQVLTYSMGVRKGDTVAVYMPMVPEAIITLLAISRIGAIHSVVFAGFSSNSLRD 230
Query 69 RLEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPS-SFVEGRD 127
R+ D S+V+IT + RGG+V+ K++VD+AL+ V+HV+++RK+ PS +F RD
Sbjct 231 RINDGDSKVVITTDESNRGGKVIETKRIVDDALRETPGVRHVLVYRKTNNPSVAFHAPRD 290
Query 128 LSGDDLLPQMRPYCPLETMDSEDML 152
L + + Y P +DSED L
Sbjct 291 LDWATEKKKYKTYYPCTPVDSEDPL 315
> xla:443879 acss2.2, MGC80104, acas2, acecs, acs, acsa, acss2;
acyl-CoA synthetase short-chain family member 2, gene 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=680
Score = 133 bits (335), Expect = 2e-31, Method: Composition-based stats.
Identities = 75/156 (48%), Positives = 97/156 (62%), Gaps = 14/156 (8%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QVC+FAN+L L V+KGD V IYLPM+PE+VYAMLACARIGAVHS+VFAGFS+ SL ER+
Sbjct 135 QVCKFANVLIKLGVKKGDRVAIYLPMIPELVYAMLACARIGAVHSIVFAGFSSESLCERI 194
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQH-----VVLFRKSAEPSSFVEG 125
D+ VLITA RG + + LKQ+ DEAL+ C+ H VV+ ++ EG
Sbjct 195 LDSQCVVLITADGVFRGEKPINLKQIADEALQKCNEKMHHVKNCVVVKHLKGRLNNQNEG 254
Query 126 R---------DLSGDDLLPQMRPYCPLETMDSEDML 152
+ D D+L+ C E +D+ED L
Sbjct 255 KFQTAWNSDVDRWLDELMADASEECEPEWLDAEDPL 290
> pfa:PFF1350c acetyl-CoA synthetase (EC:6.2.1.1); K01895 acetyl-CoA
synthetase [EC:6.2.1.1]
Length=997
Score = 133 bits (334), Expect = 2e-31, Method: Composition-based stats.
Identities = 56/109 (51%), Positives = 85/109 (77%), Gaps = 0/109 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
++ C+ ANLLK + V+K D VTIYLPM+PE++Y+MLAC RIGA+H+VVFAG+SA SL++R
Sbjct 149 EKTCKVANLLKLIGVKKQDTVTIYLPMIPELIYSMLACVRIGAIHNVVFAGYSAASLSDR 208
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAE 118
+ D+ S VLIT+ G+RGG++ +LKQ+ D A+ +C +Q ++F+ +
Sbjct 209 IIDSRSTVLITSDFGLRGGKLTKLKQIADVAMDMCGMIQTCIVFKNKTK 257
Score = 42.7 bits (99), Expect = 5e-04, Method: Composition-based stats.
Identities = 17/29 (58%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 124 EGRDLSGDDLLPQMRPYCPLETMDSEDML 152
+GRD+ G LL MR YCP+E +DSED L
Sbjct 523 KGRDIDGTALLKNMRSYCPIEYVDSEDFL 551
> sce:YLR153C ACS2; Acetyl-coA synthetase isoform which, along
with Acs1p, is the nuclear source of acetyl-coA for histone
acetylation; mutants affect global transcription; required
for growth on glucose; expressed under anaerobic conditions
(EC:6.2.1.1); K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=683
Score = 131 bits (329), Expect = 1e-30, Method: Composition-based stats.
Identities = 68/143 (47%), Positives = 94/143 (65%), Gaps = 1/143 (0%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
+V + A +LKS V+KGD V IYLPM+PE V AMLA ARIGA+HSVVFAGFSA SL +R+
Sbjct 131 KVSQIAGVLKSWGVKKGDTVAIYLPMIPEAVIAMLAVARIGAIHSVVFAGFSAGSLKDRV 190
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSA-EPSSFVEGRDLS 129
DA+S+V+IT +G RGG+ + K++VDE L V +++F+++ E GRD
Sbjct 191 VDANSKVVITCDEGKRGGKTINTKKIVDEGLNGVDLVSRILVFQRTGTEGIPMKAGRDYW 250
Query 130 GDDLLPQMRPYCPLETMDSEDML 152
+ + R Y P + D+ED L
Sbjct 251 WHEEAAKQRTYLPPVSCDAEDPL 273
> dre:541435 acss1, MGC158738, MGC174587, cb395, fi28d10, sb:cb395,
wu:fa05a10, wu:fc49f03, wu:fi28d10, zgc:113194, zgc:158738;
acyl-CoA synthetase short-chain family member 1 (EC:6.2.1.13);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=693
Score = 129 bits (323), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 67/144 (46%), Positives = 98/144 (68%), Gaps = 3/144 (2%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+ CR AN LKS V++G+ V IY+P+ P V AMLACARIGAVH+VVFAGFS+ +LA R
Sbjct 152 EMTCRLANTLKSHGVQRGERVAIYMPVSPMAVAAMLACARIGAVHTVVFAGFSSEALAGR 211
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGR-DL 128
++DA + +IT +G+RGGRV LK VD+A+K C V+HV + +++ +S G+ D+
Sbjct 212 IQDAQCKFVITCNQGVRGGRVFDLKSTVDKAVKSCPSVRHVFVAKRTN--NSVPMGKLDI 269
Query 129 SGDDLLPQMRPYCPLETMDSEDML 152
++++ C E M+SE+ML
Sbjct 270 PLEEVMAGQSSECAPEPMESEEML 293
> ath:AT5G36880 acetyl-CoA synthetase, putative / acetate-CoA
ligase, putative (EC:6.2.1.1); K01895 acetyl-CoA synthetase
[EC:6.2.1.1]
Length=743
Score = 123 bits (309), Expect = 2e-28, Method: Composition-based stats.
Identities = 67/150 (44%), Positives = 91/150 (60%), Gaps = 8/150 (5%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
+VC+ AN LK V+KGD V IYLPM+ E+ AMLACARIGAVHSVVFAGFSA+SLA+R+
Sbjct 206 RVCQLANYLKDNGVKKGDAVVIYLPMLMELPIAMLACARIGAVHSVVFAGFSADSLAQRI 265
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCS--------CVQHVVLFRKSAEPSSF 122
D V++T RG + + LK +VD AL S C+ + + E + +
Sbjct 266 VDCKPNVILTCNAVKRGPKTINLKAIVDAALDQSSKDGVSVGICLTYDNSLATTRENTKW 325
Query 123 VEGRDLSGDDLLPQMRPYCPLETMDSEDML 152
GRD+ D++ Q C +E +D+ED L
Sbjct 326 QNGRDVWWQDVISQYPTSCEVEWVDAEDPL 355
> hsa:55902 ACSS2, ACAS2, ACECS, ACS, ACSA, DKFZp762G026, dJ1161H23.1;
acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=714
Score = 118 bits (295), Expect = 8e-27, Method: Composition-based stats.
Identities = 53/94 (56%), Positives = 73/94 (77%), Gaps = 0/94 (0%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QVC+F+N+L+ ++KGD V IY+PM+PE+V AMLACARIGA+HS+VFAGFS+ SL ER+
Sbjct 144 QVCQFSNVLRKQGIQKGDRVAIYMPMIPELVVAMLACARIGALHSIVFAGFSSESLCERI 203
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEALKLC 104
D+ +LIT RG ++V LK++ DEAL+ C
Sbjct 204 LDSSCSLLITTDAFYRGEKLVNLKELADEALQKC 237
> cel:C36A4.9 hypothetical protein; K01895 acetyl-CoA synthetase
[EC:6.2.1.1]
Length=680
Score = 115 bits (287), Expect = 7e-26, Method: Composition-based stats.
Identities = 67/156 (42%), Positives = 96/156 (61%), Gaps = 14/156 (8%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QV +F+ +L+S V++GD+V +YLPM+PE+ AMLACARIGA+HSVVFAGFSA SLA R+
Sbjct 135 QVVQFSAVLRSHGVKRGDVVALYLPMIPELAVAMLACARIGAMHSVVFAGFSAESLAARV 194
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSC----VQHVVL---FRKSAEPSSF- 122
DA RVL+TA RG + + LK + D A L S V+ +++ ++ +P
Sbjct 195 VDARCRVLVTADGVFRGAKPIGLKSIADAAAVLASQEDVKVEAIIMVEHLKRVTKPDGVE 254
Query 123 ---VEGRDLS---GDDLLPQMRPYCPLETMDSEDML 152
V+ D++ ++L P+E MDSED L
Sbjct 255 LPKVDYTDITVIYDSEMLKCAGVDSPVEWMDSEDPL 290
> mmu:60525 Acss2, 1110017C11Rik, ACAS, Acas1, Acas2, AceCS1,
Acs1; acyl-CoA synthetase short-chain family member 2 (EC:6.2.1.1);
K01895 acetyl-CoA synthetase [EC:6.2.1.1]
Length=701
Score = 112 bits (279), Expect = 6e-25, Method: Composition-based stats.
Identities = 67/168 (39%), Positives = 95/168 (56%), Gaps = 26/168 (15%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QVC+F+N+L+ ++KGD V IY+PM+ E+V AMLACAR+GA+HS+VFAGFSA SL ER+
Sbjct 144 QVCQFSNVLRKQGIQKGDRVAIYMPMILELVVAMLACARLGALHSIVFAGFSAESLCERI 203
Query 71 EDAHSRVLITATKGMRGGRVVRLKQVVDEAL----------KLCSCVQHV--VLFRKSAE 118
D+ +LIT RG ++V LK++ DE+L + C V+H+ +
Sbjct 204 LDSSCCLLITTDAFYRGEKLVNLKELADESLEKCREKGFPVRCCIVVKHLGRAELGMNDS 263
Query 119 PSS--------------FVEGRDLSGDDLLPQMRPYCPLETMDSEDML 152
PS + EG DL +L+ Q C E D+ED L
Sbjct 264 PSQSPPVKRPCPDVQICWNEGVDLWWHELMQQAGDECEPEWCDAEDPL 311
> hsa:79611 ACSS3, FLJ21963; acyl-CoA synthetase short-chain family
member 3 (EC:6.2.1.1); K01908 propionyl-CoA synthetase
[EC:6.2.1.17]
Length=686
Score = 103 bits (258), Expect = 2e-22, Method: Composition-based stats.
Identities = 51/136 (37%), Positives = 84/136 (61%), Gaps = 8/136 (5%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+QV + A +L ++KGD V IY+PM+P+ +Y MLACARIGA+HS++F GF++ L+ R
Sbjct 151 EQVSKLAGVLVKHGIKKGDTVVIYMPMIPQAMYTMLACARIGAIHSLIFGGFASKELSSR 210
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQH-----VVLFRKSAEPSSFVE 124
++ +V++TA+ G+ GR V +V+EALK+ QH ++ R + E
Sbjct 211 IDHVKPKVVVTASFGIEPGRRVEYVPLVEEALKIG---QHKPDKILIYNRPNMEAVPLAP 267
Query 125 GRDLSGDDLLPQMRPY 140
GRDL D+ + + + +
Sbjct 268 GRDLDWDEEMAKAQSH 283
> mmu:380660 Acss3, 8430416H19Rik, Gm874; acyl-CoA synthetase
short-chain family member 3 (EC:6.2.1.1); K01908 propionyl-CoA
synthetase [EC:6.2.1.17]
Length=682
Score = 103 bits (258), Expect = 2e-22, Method: Composition-based stats.
Identities = 50/136 (36%), Positives = 86/136 (63%), Gaps = 8/136 (5%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+QV + A +L V+KGD V IY+PM+P+ +Y MLACARIGA+HS++F GF++ L+ R
Sbjct 146 EQVSKLAGVLVKQGVKKGDTVVIYMPMIPQAIYTMLACARIGAIHSLIFGGFASKELSTR 205
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQH-----VVLFRKSAEPSSFVE 124
++ A +V++TA+ G+ GR V +++EAL++ QH ++ R + E +
Sbjct 206 IDHAKPKVVVTASFGIEPGRKVEYIPLLEEALRIG---QHRPDRVLIYSRPNMEKVPLMS 262
Query 125 GRDLSGDDLLPQMRPY 140
GRDL ++ + + + +
Sbjct 263 GRDLDWEEEMAKAQSH 278
> eco:b0335 prpE, ECK0332, JW0326, yahU; propionate--CoA ligase;
K01908 propionyl-CoA synthetase [EC:6.2.1.17]
Length=628
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 49/120 (40%), Positives = 83/120 (69%), Gaps = 1/120 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+V A++L+SL V++GD V +Y+PM+ E +LACARIGA+HSVVF GF+++S+A R
Sbjct 91 DEVNAVASMLRSLGVQRGDRVLVYMPMIAEAHITLLACARIGAIHSVVFGGFASHSVAAR 150
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSC-VQHVVLFRKSAEPSSFVEGRDL 128
++DA ++++A G RGG+++ K+++D+A+ +HV+L + + V GRD+
Sbjct 151 IDDAKPVLIVSADAGARGGKIIPYKKLLDDAISQAQHQPRHVLLVDRGLAKMARVSGRDV 210
> dre:566019 acss3, si:dkey-231a18.1; acyl-CoA synthetase short-chain
family member 3; K01908 propionyl-CoA synthetase [EC:6.2.1.17]
Length=714
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/133 (39%), Positives = 83/133 (62%), Gaps = 2/133 (1%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
+QV R A +L V+ GD+V IY+PM+P+ ++ MLACARIGA HS++F GF++ L+ R
Sbjct 173 EQVSRLAGVLVKHGVKMGDLVVIYMPMVPQAMFTMLACARIGATHSLIFGGFASKELSSR 232
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLF--RKSAEPSSFVEGRD 127
++ A ++L+TA+ G+ GR+V +V++AL+L + H VL R S E S
Sbjct 233 IDHAKPKLLVTASFGIEPGRLVEYIPLVEKALELSTHKPHKVLIYNRPSMEKVSTRPDLT 292
Query 128 LSGDDLLPQMRPY 140
L ++ + RP+
Sbjct 293 LDWEEEMSTARPH 305
> tgo:TGME49_032580 acetyl-coenzyme A synthetase, putative (EC:6.2.1.1)
Length=821
Score = 88.2 bits (217), Expect = 1e-17, Method: Composition-based stats.
Identities = 46/139 (33%), Positives = 77/139 (55%), Gaps = 0/139 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
++V A+ L+ V +GD V IY+P++PE +MLACARIGAVH+V F GF L+ R
Sbjct 234 ERVSALADALRKAGVHEGDRVLIYMPVVPEAAISMLACARIGAVHAVTFGGFGTQELSTR 293
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLS 129
++ A + +I A+ G+ R+V K+ ++ AL+L S + + ++ +E D+
Sbjct 294 IDHAKPKAIIAASCGIEPRRIVPYKEGLEHALQLSSHKPEFKVILQRSQHECTLESSDID 353
Query 130 GDDLLPQMRPYCPLETMDS 148
D + C + +DS
Sbjct 354 WDCFIKSGSGQCDILPVDS 372
> dre:560020 hypothetical LOC560020
Length=479
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 47/142 (33%), Positives = 70/142 (49%), Gaps = 29/142 (20%)
Query 26 KGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHSRVLITATKGM 85
+ DI T+++ ++V V FAGFS+ SL ER+ DA +L+TA
Sbjct 144 QADITTLFISRNSDLV--------------VQFAGFSSESLCERIMDAQCNILVTADGVY 189
Query 86 RGGRVVRLKQVVDEALKLC-----SCVQHVVL-----FRKSAEPSSFVEGR-----DLSG 130
RG +++ LK++ EAL C S VQ V+ F S++ SS ++ D+
Sbjct 190 RGDKLINLKKIASEALDRCKESSASSVQKCVVVKHPAFITSSDASSSLQTPWDELCDIWW 249
Query 131 DDLLPQMRPYCPLETMDSEDML 152
DDL+ + C E MDSED L
Sbjct 250 DDLMDGVSDECEPEWMDSEDPL 271
> mmu:78894 Aacs, 2210408B16Rik, SUR5; acetoacetyl-CoA synthetase
(EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=672
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 51/106 (48%), Gaps = 8/106 (7%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QV FA ++ + V+KGD V YLP V AMLA A IGA+ S F N + +R
Sbjct 137 QVALFAAAMRKMGVKKGDRVVGYLPNSAHAVEAMLAAASIGAIWSSTSPDFGVNGVLDRF 196
Query 71 EDAHSRVLITAT----KGMRGGRVVRLKQVVDEALKLCSCVQHVVL 112
+++ + G G + +L++VV K +Q VVL
Sbjct 197 SQIQPKLIFSVEAVVYNGKEHGHLEKLQRVV----KGLPDLQRVVL 238
> hsa:65985 AACS, ACSF1, FLJ12389, FLJ41251, SUR-5; acetoacetyl-CoA
synthetase (EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase
[EC:6.2.1.16]
Length=672
Score = 48.9 bits (115), Expect = 7e-06, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 46/91 (50%), Gaps = 4/91 (4%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
+V FA ++ + V+KGD V YLP V AMLA A IGA+ S F N + +R
Sbjct 137 EVALFAAAMRKMGVKKGDRVVGYLPNSEHAVEAMLAAASIGAIWSSTSPDFGVNGVLDRF 196
Query 71 EDAHSRVLITATKGMRGGR----VVRLKQVV 97
+++ + + G+ + +L+QVV
Sbjct 197 SQIQPKLIFSVEAVVYNGKEHNHMEKLQQVV 227
> ath:AT1G65060 4CL3; 4CL3; 4-coumarate-CoA ligase (EC:6.2.1.12);
K01904 4-coumarate--CoA ligase [EC:6.2.1.12]
Length=561
Score = 44.7 bits (104), Expect = 1e-04, Method: Composition-based stats.
Identities = 36/138 (26%), Positives = 68/138 (49%), Gaps = 16/138 (11%)
Query 4 FAFAFSDQVCR-FANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFS 62
+ + + +CR A+ L L +RKGD++ I L E V++ + + IGAV + ++
Sbjct 76 YTYGETHLICRRVASGLYKLGIRKGDVIMILLQNSAEFVFSFMGASMIGAVSTTANPFYT 135
Query 63 ANSLAERLEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCS--------CVQHVVLFR 114
+ L ++L+ + ++++IT ++ V +LK + E L L + C+ L
Sbjct 136 SQELYKQLKSSGAKLIITHSQ-----YVDKLKN-LGENLTLITTDEPTPENCLPFSTLIT 189
Query 115 KSAEPSSFVEGRDLSGDD 132
E + F E D+ GDD
Sbjct 190 DD-ETNPFQETVDIGGDD 206
> ath:AT1G68270 AMP-dependent synthetase and ligase family protein
Length=535
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 35/138 (25%), Positives = 55/138 (39%), Gaps = 13/138 (9%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+ CR A L SL++ K D+V++ P P I A GAV + + A S+
Sbjct 47 DRCCRLAASLISLNIAKNDVVSVVAPNTPAIYEMHFAVPMAGAVLNPINTRLDATSITTI 106
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSC----VQHVVLFRKSAEPSSFVEG 125
L A ++L + + E L L SC + +V+F + V
Sbjct 107 LRHAQPKILFIHRN---------FEPLAREILHLLSCDDLQLNLLVIFIDEYNSAKRVSS 157
Query 126 RDLSGDDLLPQMRPYCPL 143
+L + L+ P PL
Sbjct 158 EELDYESLIQMGEPTSPL 175
> eco:b0037 caiC, ECK0038, JW0036, yaaM; probable crotonobetaine/carnitine-CoA
ligase (EC:6.3.2.-); K02182 crotonobetaine/carnitine-CoA
ligase [EC:6.2.1.-]
Length=517
Score = 44.3 bits (103), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 42/76 (55%), Gaps = 0/76 (0%)
Query 8 FSDQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLA 67
+ ++ R ANL +L +RKGD V ++L PE ++ A+IGA+ + A A
Sbjct 43 LNQEINRTANLFYTLGIRKGDKVALHLDNCPEFIFCWFGLAKIGAIMVPINARLLCEESA 102
Query 68 ERLEDAHSRVLITATK 83
L+++ + +L+T+ +
Sbjct 103 WILQNSQACLLVTSAQ 118
> dre:393984 aacs, MGC56105, zgc:56105; acetoacetyl-CoA synthetase
(EC:6.2.1.16); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=659
Score = 42.0 bits (97), Expect = 7e-04, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 0/78 (0%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
V FA ++ + ++ GD V YLP V AMLA A IGA+ S F N + +R+
Sbjct 138 VALFAAAMRKMGIKIGDRVVGYLPNGIHAVEAMLAAASIGAIWSSTSPDFGVNGVLDRIS 197
Query 72 DAHSRVLITATKGMRGGR 89
+++ + + G+
Sbjct 198 QIQPKLIFSVAAVVYNGK 215
> ath:AT4G05160 4-coumarate--CoA ligase, putative / 4-coumaroyl-CoA
synthase, putative; K01904 4-coumarate--CoA ligase [EC:6.2.1.12]
Length=544
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 0/72 (0%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
V R A+ L +RK D+V I+ P + LA IG V + ++ N ++++++
Sbjct 64 VARLAHGFHRLGIRKNDVVLIFAPNSYQFPLCFLAVTAIGGVFTTANPLYTVNEVSKQIK 123
Query 72 DAHSRVLITATK 83
D++ +++I+ +
Sbjct 124 DSNPKIIISVNQ 135
> ath:AT1G55320 AAE18; AAE18 (ACYL-ACTIVATING ENZYME 18); catalytic/
ligase
Length=511
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 32/115 (27%), Positives = 53/115 (46%), Gaps = 4/115 (3%)
Query 26 KGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHSRVLITATKGM 85
KGD + I +PM + V LA G + + F+A +A RL+ + ++ + T +
Sbjct 13 KGDTIAIDMPMTVDAVIIYLAIILAGCIVVSIADSFAAKEIATRLKISKAKGIFTQDYIL 72
Query 86 RGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVEGRDLSGDDLLPQMRPY 140
RGGR L V EA + + +VL E + +D+S D L +P+
Sbjct 73 RGGRRFPLYSRVVEA----APSKVIVLPASGTELHVQLREQDVSWMDFLSNAKPH 123
> cel:F28D1.9 hypothetical protein; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=684
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 37/74 (50%), Gaps = 0/74 (0%)
Query 8 FSDQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLA 67
F+ R+AN + L R GD+V +Y+ E V A + A+IG V + + + L
Sbjct 142 FNAHCNRYANYFQGLGYRSGDVVALYMENSVEFVAAWMGLAKIGVVTAWINSNLKREQLV 201
Query 68 ERLEDAHSRVLITA 81
+ + ++ +IT+
Sbjct 202 HCITASKTKAIITS 215
> eco:b0586 entF, ECK0579, JW0578; enterobactin synthase multienzyme
complex component, ATP-dependent (EC:2.7.7.-); K02364
enterobactin synthetase component F [EC:2.7.7.-]
Length=1293
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 39/79 (49%), Gaps = 1/79 (1%)
Query 3 LFAF-AFSDQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGF 61
LF++ +QV ANLL+ V+ GD V + LP + A+ A GA + G+
Sbjct 480 LFSYREMREQVVALANLLRERGVKPGDSVAVALPRSVFLTLALHAIVEAGAAWLPLDTGY 539
Query 62 SANSLAERLEDAHSRVLIT 80
+ L LEDA +LIT
Sbjct 540 PDDRLKMMLEDARPSLLIT 558
> ath:AT1G65880 BZO1; BZO1 (BENZOYLOXYGLUCOSINOLATE 1); benzoate-CoA
ligase
Length=580
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 0/69 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+ CR A L SL++ K D+V++ P P + A GAV + + A S+A
Sbjct 47 DRCCRLAASLISLNISKNDVVSVMAPNTPALYEMHFAVPMAGAVLNPINTRLDATSIAAI 106
Query 70 LEDAHSRVL 78
L A ++L
Sbjct 107 LRHAKPKIL 115
> ath:AT3G21240 4CL2; 4CL2 (4-COUMARATE:COA LIGASE 2); 4-coumarate-CoA
ligase (EC:6.2.1.12); K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=556
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/70 (27%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 14 RFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDA 73
+ A L +L V++ D+V I LP PE+V LA + IGA+ + F+ ++++ + +
Sbjct 75 KLAAGLHNLGVKQHDVVMILLPNSPEVVLTFLAASFIGAITTSANPFFTPAEISKQAKAS 134
Query 74 HSRVLITATK 83
+++++T ++
Sbjct 135 AAKLIVTQSR 144
> ath:AT1G65890 AAE12; AAE12 (ACYL ACTIVATING ENZYME 12); catalytic
Length=578
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 33/116 (28%), Positives = 51/116 (43%), Gaps = 3/116 (2%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+ CR A L SL++ K D+V++ P P + A GAV + + A S+A
Sbjct 47 DRCCRLAASLISLNIGKNDVVSVVAPNTPAMYEMHFAVPMAGAVLNPINTRLDATSIAAI 106
Query 70 LEDAHSRVLITATKGMRGGR-VVRLKQVVDEALKLCSCVQHVVLF--RKSAEPSSF 122
L A ++L R +++L D L L H + F R S+E S +
Sbjct 107 LRHAKPKILFIYRSFEPLAREILQLLSSEDSNLNLPVIFIHEIDFPKRVSSEESDY 162
> cel:F41C3.3 acs-11; fatty Acid CoA Synthetase family member
(acs-11)
Length=505
Score = 39.3 bits (90), Expect = 0.005, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 56/111 (50%), Gaps = 11/111 (9%)
Query 18 LLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHSRV 77
L + +++KGD V + + +AC +IGA++ V G++ + A ++DA +
Sbjct 63 LTEKYNIKKGDRVMARVSKTTDTAALYIACLQIGALYIPVNPGYTESEAAHYIKDATPSI 122
Query 78 LITATKGMRGG-----RVVRLKQVVDEA--LKLCSCVQHVVLFRKSAEPSS 121
L++ + + RV+ ++ EA L C+ ++HV + ++P+S
Sbjct 123 LVSCNEELDKVFRDKIRVINEDKLASEAGSLNACTMIEHV----EKSDPAS 169
> mmu:20216 Acsm3, Sa, Sah; acyl-CoA synthetase medium-chain family
member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA synthetase
[EC:6.2.1.2]
Length=580
Score = 38.9 bits (89), Expect = 0.007, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 14 RFANLL-KSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLED 72
+FAN+L ++ +++GD V + LP +PE A +AC R G V + + RL+
Sbjct 100 KFANILTEACSLQRGDRVMVILPKIPEWWLANVACLRTGTVLIPGTTQLTQKDILYRLQS 159
Query 73 AHSRVLIT 80
+ ++ +IT
Sbjct 160 SKAKCIIT 167
> ath:AT1G51680 4CL1; 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumarate-CoA
ligase (EC:6.2.1.12); K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=539
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 63/130 (48%), Gaps = 15/130 (11%)
Query 16 ANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHS 75
AN K L V + D+V + LP PE V + LA + GA + F+ +A++ + +++
Sbjct 80 ANFHK-LGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTPAEIAKQAKASNT 138
Query 76 RVLITATKGMRGGRVVRLKQVVDEALKLC-----------SCVQHVVLFRKSAEPSSFVE 124
+++IT + + ++ Q D + +C C++ L + + E S ++
Sbjct 139 KLIITEARYVDK---IKPLQNDDGVVIVCIDDNESVPIPEGCLRFTELTQSTTEASEVID 195
Query 125 GRDLSGDDLL 134
++S DD++
Sbjct 196 SVEISPDDVV 205
> ath:AT1G66120 acyl-activating enzyme 11 (AAE11)
Length=572
Score = 37.7 bits (86), Expect = 0.013, Method: Composition-based stats.
Identities = 20/69 (28%), Positives = 35/69 (50%), Gaps = 0/69 (0%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+ CR A L SL++ + D+V+I P +P + + GAV + + A ++A
Sbjct 47 DRCCRLAASLLSLNITRNDVVSILAPNVPAMYEMHFSVPMTGAVLNPINTRLDAKTIAII 106
Query 70 LEDAHSRVL 78
L A ++L
Sbjct 107 LRHAEPKIL 115
> cel:K03A1.5 sur-5; SUppressor of activated let-60 Ras family
member (sur-5); K01907 acetoacetyl-CoA synthetase [EC:6.2.1.16]
Length=700
Score = 37.4 bits (85), Expect = 0.020, Method: Composition-based stats.
Identities = 26/111 (23%), Positives = 46/111 (41%), Gaps = 6/111 (5%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
V R A L++ + GD V ++P + + A+ A A +GA F + +R
Sbjct 166 VYRIATSLRNYGIGPGDTVCGFVPNTYDTLVAVFATAAVGAAWCSASVDFGPAGVLDRFR 225
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSF 122
H +VL T V K+++D+ K+ V+ + K +F
Sbjct 226 QVHPKVLFTVN------HVTYKKKLIDQTDKINEIVKELPTLEKIVVSDTF 270
> hsa:6296 ACSM3, SA, SAH; acyl-CoA synthetase medium-chain family
member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA synthetase
[EC:6.2.1.2]
Length=586
Score = 37.0 bits (84), Expect = 0.023, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 37/68 (54%), Gaps = 1/68 (1%)
Query 14 RFANLL-KSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLED 72
+FAN+L ++ +++GD V + LP +PE A +AC R G V + + RL+
Sbjct 106 KFANILSEACSLQRGDRVILILPRVPEWWLANVACLRTGTVLIPGTTQLTQKDILYRLQS 165
Query 73 AHSRVLIT 80
+ + +IT
Sbjct 166 SKANCIIT 173
> cel:AH10.1 hypothetical protein; K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=566
Score = 36.6 bits (83), Expect = 0.032, Method: Composition-based stats.
Identities = 28/113 (24%), Positives = 49/113 (43%), Gaps = 9/113 (7%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
V A L L + GD+ P PE + AMLA + G S A F+ L + +
Sbjct 57 VNSLATALVKLGFKPGDVAAQAFPNCPEFLIAMLAVMKCGGAMSNASAIFTDYELQLQFK 116
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVE 124
D+++ ++ T R+ R+++ V + C V+ ++ R + F E
Sbjct 117 DSNTSIVFTDED-----RLARVRRSVAK----CPGVRKIICLRTFPLRAEFPE 160
> tgo:TGME49_019230 acetoacetyl-CoA synthetase, putative (EC:6.2.1.16)
Length=535
Score = 36.6 bits (83), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 0/81 (0%)
Query 11 QVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERL 70
QV + ++KGD V + PE V MLA +G + S F+ L R
Sbjct 9 QVAVMMQFFRRTGLQKGDRVAAVVCNTPETVVTMLAVTGLGGIWSSCSPDFTLPVLHSRF 68
Query 71 EDAHSRVLITATKGMRGGRVV 91
D RVL+T+ GR
Sbjct 69 GDLSPRVLVTSDVYQLKGRTT 89
> ath:AT3G21230 4CL5; 4CL5 (4-coumarate:CoA ligase 5); 4-coumarate-CoA
ligase (EC:6.2.1.12); K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=570
Score = 35.4 bits (80), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 14 RFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSV 56
R A + L +R GD+V + LP PE + LA A +GAV +
Sbjct 85 RIAAGIHRLGIRHGDVVMLLLPNSPEFALSFLAVAYLGAVSTT 127
> mmu:26457 Slc27a1, FATP1, Fatp; solute carrier family 27 (fatty
acid transporter), member 1; K08745 solute carrier family
27 (fatty acid transporter), member 1/4 [EC:6.2.1.-]
Length=646
Score = 35.4 bits (80), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 40/90 (44%), Gaps = 1/90 (1%)
Query 16 ANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLEDAHS 75
ANL + L GD+V ++L PE V L A+ G V +++ LA L + +
Sbjct 117 ANLFRQLGFAPGDVVAVFLEGRPEFVGLWLGLAKAGVVAALLNVNLRREPLAFCLGTSAA 176
Query 76 RVLITATKGMRGGRVVRLKQVVDEALKLCS 105
+ LI + M +Q+ LK CS
Sbjct 177 KALIYGGE-MAAAVAEVSEQLGKSLLKFCS 205
> cel:ZK1127.2 hypothetical protein; K01904 4-coumarate--CoA ligase
[EC:6.2.1.12]
Length=565
Score = 35.0 bits (79), Expect = 0.086, Method: Composition-based stats.
Identities = 28/113 (24%), Positives = 48/113 (42%), Gaps = 9/113 (7%)
Query 12 VCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLE 71
V A L L + GD+ + P PE + AMLA + G S A F+ L +
Sbjct 57 VNSLATALVKLGFKPGDVASQAFPNCPEFLVAMLAVMKCGGAMSNASAIFTDYELQLQFC 116
Query 72 DAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEPSSFVE 124
D+++ ++ T R+ R+++ A C V+ ++ R + F E
Sbjct 117 DSNTSIVFTDED-----RLARIRR----ATAKCPGVRKIICLRTFPLRTEFPE 160
> ath:AT1G76290 AMP-dependent synthetase and ligase family protein
Length=546
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 32/144 (22%), Positives = 58/144 (40%), Gaps = 18/144 (12%)
Query 10 DQVCRFANLLKSLDVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
D+ R A+ L L + + D+V P +P + GAV V+ F + LA
Sbjct 47 DRCVRLASALSDLGLSRHDVVAALAPNVPALCELYFGAPMAGAVLCVLNTTFDSQMLAMA 106
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQH----VVLFRKSAEPSSFVEG 125
LE +V ++ + V +E+L L S ++ + + E S + +
Sbjct 107 LEKTKPKVFFVDSEFL---------SVAEESLSLLSNIEEKPLIITITENPTEQSKYEQY 157
Query 126 RDL--SGDDLLPQMRPYCPLETMD 147
D +G+ P +P P++ D
Sbjct 158 EDFLSTGN---PNFKPIRPVDECD 178
> mmu:117147 Acsm1, Acas3, Bucs1, Macs; acyl-CoA synthetase medium-chain
family member 1 (EC:6.2.1.2); K01896 medium-chain
acyl-CoA synthetase [EC:6.2.1.2]
Length=573
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 17/72 (23%), Positives = 36/72 (50%), Gaps = 1/72 (1%)
Query 10 DQVCRFANLLKSL-DVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAE 68
D CR AN+ + + +++GD + + LP +PE + C R G + A +
Sbjct 89 DLTCRTANVFEQICGLQQGDHLALILPRVPEWWLVTVGCMRTGIIFMPGTTQLKAKDILY 148
Query 69 RLEDAHSRVLIT 80
R++ + ++ ++T
Sbjct 149 RIQISRAKAIVT 160
> mmu:272428 Acsm5, C730019D22, C730027J19Rik; acyl-CoA synthetase
medium-chain family member 5 (EC:6.2.1.2); K01896 medium-chain
acyl-CoA synthetase [EC:6.2.1.2]
Length=578
Score = 34.7 bits (78), Expect = 0.12, Method: Composition-based stats.
Identities = 27/110 (24%), Positives = 50/110 (45%), Gaps = 10/110 (9%)
Query 11 QVCRFANLLKSL-DVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAER 69
Q + AN+L+ + ++ GD + + LP +P+ +AC R G V + +A L R
Sbjct 97 QSRKAANVLEGVCGLQPGDRMMLVLPRLPDWWLISVACMRTGVVMIPGVSQLTAKDLKYR 156
Query 70 LEDAHSRVLITATKGMRGGRVVRLKQVVDEALKLCSCVQHVVLFRKSAEP 119
L+ A ++ ++T+ L VD C +Q +L ++ P
Sbjct 157 LQAARAKSIVTSDA---------LAPQVDAISADCPSLQTKLLVSDTSRP 197
> dre:100002035 acsm3, zgc:172040; acyl-CoA synthetase medium-chain
family member 3 (EC:6.2.1.2); K01896 medium-chain acyl-CoA
synthetase [EC:6.2.1.2]
Length=591
Score = 33.9 bits (76), Expect = 0.23, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Query 14 RFANLLKSL-DVRKGDIVTIYLPMMPEIVYAMLACARIGAVHSVVFAGFSANSLAERLED 72
+ AN+L+ + + +GD V + LP +PE +AC R G V + +A + RL
Sbjct 109 KLANVLRQVCSLERGDRVFLILPRVPEWWLVNVACIRTGTVLIPGTSQLTARDMRHRLIT 168
Query 73 AHSRVLIT 80
+ ++ +IT
Sbjct 169 SGAKCVIT 176
Lambda K H
0.326 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3264639800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40