bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2822_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
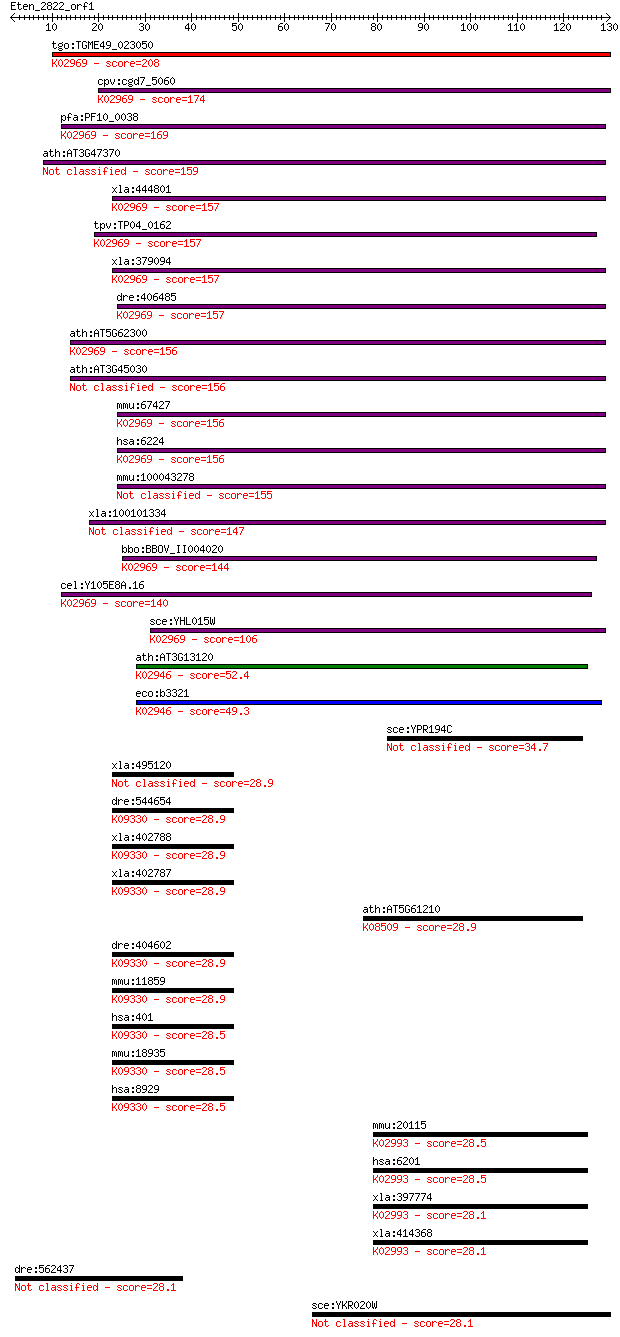
tgo:TGME49_023050 40s ribosomal protein S20, putative ; K02969... 208 3e-54
cpv:cgd7_5060 40S ribosomal protein S20 ; K02969 small subunit... 174 8e-44
pfa:PF10_0038 40S ribosomal protein S20e, putative; K02969 sma... 169 3e-42
ath:AT3G47370 40S ribosomal protein S20 (RPS20B) 159 2e-39
xla:444801 MGC82136 protein; K02969 small subunit ribosomal pr... 157 7e-39
tpv:TP04_0162 40S ribosomal protein S20; K02969 small subunit ... 157 1e-38
xla:379094 rps20, MGC52591; ribosomal protein S20; K02969 smal... 157 1e-38
dre:406485 rps20, wu:fb81b03, zgc:77758; ribosomal protein S20... 157 1e-38
ath:AT5G62300 40S ribosomal protein S20 (RPS20C); K02969 small... 156 1e-38
ath:AT3G45030 40S ribosomal protein S20 (RPS20A) 156 1e-38
mmu:67427 Rps20, 4632426K06Rik, Dsk4, MGC102408; ribosomal pro... 156 2e-38
hsa:6224 RPS20, FLJ27451, MGC102930; ribosomal protein S20; K0... 156 2e-38
mmu:100043278 Gm4332; predicted gene 4332 155 2e-38
xla:100101334 hypothetical protein LOC100101334 147 1e-35
bbo:BBOV_II004020 18.m06332; 40S ribosomal protein S20; K02969... 144 7e-35
cel:Y105E8A.16 rps-20; Ribosomal Protein, Small subunit family... 140 1e-33
sce:YHL015W RPS20, URP2; Rps20p; K02969 small subunit ribosoma... 106 1e-23
ath:AT3G13120 30S ribosomal protein S10, chloroplast, putative... 52.4 4e-07
eco:b3321 rpsJ, ECK3308, JW3283, nusE; 30S ribosomal subunit p... 49.3 3e-06
sce:YPR194C OPT2; Oligopeptide transporter; member of the OPT ... 34.7 0.086
xla:495120 phox2b, phox2, xphox2b; paired-like homeobox 2b 28.9 3.7
dre:544654 phox2b; paired-like homeobox 2b; K09330 paired meso... 28.9 3.9
xla:402788 phox2a, phox2, xphox2a; paired-like homeobox 2a; K0... 28.9 3.9
xla:402787 phox2b; paired-like homeobox 2b; K09330 paired meso... 28.9 3.9
ath:AT5G61210 SNAP33; SNAP33 (SOLUBLE N-ETHYLMALEIMIDE-SENSITI... 28.9 4.2
dre:404602 phox2a, MGC77364, arix, sou, zgc:77364; paired-like... 28.9 4.3
mmu:11859 Phox2a, Arix, Phox2, Pmx2, Pmx2a; paired-like homeob... 28.9 4.7
hsa:401 PHOX2A, ARIX, CFEOM2, FEOM2, MGC52227, NCAM2, PMX2A; p... 28.5 5.6
mmu:18935 Phox2b, Dilp1, NBPhox, Pmx2b; paired-like homeobox 2... 28.5 5.7
hsa:8929 PHOX2B, NBLST2, NBPhox, PMX2B; paired-like homeobox 2... 28.5 5.7
mmu:20115 Rps7, Rps7A, S7; ribosomal protein S7 (EC:3.6.5.3); ... 28.5 6.2
hsa:6201 RPS7, DBA8; ribosomal protein S7 (EC:3.6.5.3); K02993... 28.5 6.2
xla:397774 rps7, rpS8A; ribosomal protein S8 (EC:3.6.5.3); K02... 28.1 6.7
xla:414368 rps7, MGC53555, dba8, rpS8B; ribosomal protein S7; ... 28.1 6.7
dre:562437 ubfd1, MGC162578, im:6905175, zgc:162578; ubiquitin... 28.1 6.8
sce:YKR020W VPS51, API3, VPS67, WHI6; Component of the GARP (G... 28.1 7.9
> tgo:TGME49_023050 40s ribosomal protein S20, putative ; K02969
small subunit ribosomal protein S20e
Length=233
Score = 208 bits (530), Expect = 3e-54, Method: Compositional matrix adjust.
Identities = 100/120 (83%), Positives = 110/120 (91%), Gaps = 0/120 (0%)
Query 10 ASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVK 69
A MSK +K GLEGEEQRLHRIRITLTSKDLKSIERVC+ELI AK++ LQ GPVRLPVK
Sbjct 114 AKMSKLMKGGLEGEEQRLHRIRITLTSKDLKSIERVCTELINGAKEKNLQVAGPVRLPVK 173
Query 70 TLRVTTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSNI 129
TLR+TTRKSPCGEGTNT+DRFELRIYKRLIDLHS SDVV+QITSINIDPGVEVEVT+ ++
Sbjct 174 TLRITTRKSPCGEGTNTWDRFELRIYKRLIDLHSPSDVVKQITSINIDPGVEVEVTIIDM 233
> cpv:cgd7_5060 40S ribosomal protein S20 ; K02969 small subunit
ribosomal protein S20e
Length=135
Score = 174 bits (440), Expect = 8e-44, Method: Compositional matrix adjust.
Identities = 80/112 (71%), Positives = 102/112 (91%), Gaps = 2/112 (1%)
Query 20 LEGEEQR--LHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRK 77
GE+Q+ LH+IRITL+SK+LKSIERVC++LI +AKQ+ L+ GPVR+PVKTLR+TTRK
Sbjct 24 FNGEQQQAPLHKIRITLSSKNLKSIERVCADLIQSAKQKDLKVTGPVRMPVKTLRITTRK 83
Query 78 SPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSNI 129
SPCGEG+ T+DR+E+RIYKR+IDL+S+SDVV+QITSINID GVEVEVT+S+I
Sbjct 84 SPCGEGSKTWDRYEMRIYKRVIDLYSSSDVVKQITSINIDAGVEVEVTISDI 135
> pfa:PF10_0038 40S ribosomal protein S20e, putative; K02969 small
subunit ribosomal protein S20e
Length=118
Score = 169 bits (427), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 78/117 (66%), Positives = 100/117 (85%), Gaps = 0/117 (0%)
Query 12 MSKALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTL 71
MSK +K ++ E+ RL RIRI LTSK+L++IE+VCS+++ AK++ L +GPVRLPVKTL
Sbjct 1 MSKLMKGAIDNEKYRLRRIRIALTSKNLRAIEKVCSDIMKGAKEKNLNVSGPVRLPVKTL 60
Query 72 RVTTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
R+TTRKSPCGEGTNT+DRFELRIYKRLIDL+S +VV Q+TSINIDP VEVEV +++
Sbjct 61 RITTRKSPCGEGTNTWDRFELRIYKRLIDLYSQCEVVTQMTSINIDPVVEVEVIITD 117
> ath:AT3G47370 40S ribosomal protein S20 (RPS20B)
Length=122
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 70/121 (57%), Positives = 101/121 (83%), Gaps = 0/121 (0%)
Query 8 LAASMSKALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLP 67
+A K K GLE +++H+IRITL+SK++K++E+VC++L+ AK ++L+ GPVR+P
Sbjct 1 MAYEPMKPTKAGLEAPLEQIHKIRITLSSKNVKNLEKVCTDLVRGAKDKRLRVKGPVRMP 60
Query 68 VKTLRVTTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVS 127
K L++TTRK+PCGEGTNT+DRFELR++KR+IDL S+ DVV+QITSI I+PGVEVEVT++
Sbjct 61 TKVLKITTRKAPCGEGTNTWDRFELRVHKRVIDLFSSPDVVKQITSITIEPGVEVEVTIA 120
Query 128 N 128
+
Sbjct 121 D 121
> xla:444801 MGC82136 protein; K02969 small subunit ribosomal
protein S20e
Length=119
Score = 157 bits (397), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 68/106 (64%), Positives = 96/106 (90%), Gaps = 0/106 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGE 82
+E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGE
Sbjct 13 QEVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKNLKVKGPVRMPTKTLRITTRKTPCGE 72
Query 83 GTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
G+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 73 GSKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 118
> tpv:TP04_0162 40S ribosomal protein S20; K02969 small subunit
ribosomal protein S20e
Length=116
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 71/108 (65%), Positives = 93/108 (86%), Gaps = 0/108 (0%)
Query 19 GLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKS 78
G E+ R+H+IR+TL S+DLKSIE+ C ++I AK++ L+ GPVR+PV+ L++TTRKS
Sbjct 9 GEYDEDNRIHKIRLTLVSRDLKSIEKACRDIITGAKEKNLRVIGPVRMPVRKLKLTTRKS 68
Query 79 PCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTV 126
PCGEGTNT+DRFE+RIYKRLIDLHS+S VVR+ITSI +DPGVEVEV++
Sbjct 69 PCGEGTNTWDRFEMRIYKRLIDLHSSSAVVRKITSIPLDPGVEVEVSI 116
> xla:379094 rps20, MGC52591; ribosomal protein S20; K02969 small
subunit ribosomal protein S20e
Length=119
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 68/106 (64%), Positives = 96/106 (90%), Gaps = 0/106 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGE 82
+E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGE
Sbjct 13 QEVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKNLKVKGPVRMPTKTLRITTRKTPCGE 72
Query 83 GTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
G+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 73 GSKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 118
> dre:406485 rps20, wu:fb81b03, zgc:77758; ribosomal protein S20;
K02969 small subunit ribosomal protein S20e
Length=119
Score = 157 bits (396), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 95/105 (90%), Gaps = 0/105 (0%)
Query 24 EQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEG 83
E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGEG
Sbjct 14 EVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKSLKVKGPVRMPTKTLRITTRKTPCGEG 73
Query 84 TNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 74 SKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 118
> ath:AT5G62300 40S ribosomal protein S20 (RPS20C); K02969 small
subunit ribosomal protein S20e
Length=124
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 69/115 (60%), Positives = 99/115 (86%), Gaps = 0/115 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRV 73
K K GLE +++H+IRITL+SK++K++E+VC++L+ AK ++L+ GPVR+P K L++
Sbjct 9 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEKVCTDLVRGAKDKRLRVKGPVRMPTKVLKI 68
Query 74 TTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
TTRK+PCGEGTNT+DRFELR++KR+IDL S+ DVV+QITSI I+PGVEVEVT+++
Sbjct 69 TTRKAPCGEGTNTWDRFELRVHKRVIDLFSSPDVVKQITSITIEPGVEVEVTIAD 123
> ath:AT3G45030 40S ribosomal protein S20 (RPS20A)
Length=124
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 69/115 (60%), Positives = 99/115 (86%), Gaps = 0/115 (0%)
Query 14 KALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRV 73
K K GLE +++H+IRITL+SK++K++E+VC++L+ AK ++L+ GPVR+P K L++
Sbjct 9 KPGKAGLEEPLEQIHKIRITLSSKNVKNLEKVCTDLVRGAKDKRLRVKGPVRMPTKVLKI 68
Query 74 TTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
TTRK+PCGEGTNT+DRFELR++KR+IDL S+ DVV+QITSI I+PGVEVEVT+++
Sbjct 69 TTRKAPCGEGTNTWDRFELRVHKRVIDLFSSPDVVKQITSITIEPGVEVEVTIAD 123
> mmu:67427 Rps20, 4632426K06Rik, Dsk4, MGC102408; ribosomal protein
S20; K02969 small subunit ribosomal protein S20e
Length=119
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 95/105 (90%), Gaps = 0/105 (0%)
Query 24 EQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEG 83
E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGEG
Sbjct 14 EVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKNLKVKGPVRMPTKTLRITTRKTPCGEG 73
Query 84 TNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 74 SKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 118
> hsa:6224 RPS20, FLJ27451, MGC102930; ribosomal protein S20;
K02969 small subunit ribosomal protein S20e
Length=119
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 95/105 (90%), Gaps = 0/105 (0%)
Query 24 EQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEG 83
E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGEG
Sbjct 14 EVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKNLKVKGPVRMPTKTLRITTRKTPCGEG 73
Query 84 TNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 74 SKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 118
> mmu:100043278 Gm4332; predicted gene 4332
Length=115
Score = 155 bits (393), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 95/105 (90%), Gaps = 0/105 (0%)
Query 24 EQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEG 83
E +HRIRITLTS+++KS+E+VC++LI AK++ L+ GPVR+P KTLR+TTRK+PCGEG
Sbjct 10 EVAIHRIRITLTSRNVKSLEKVCADLIRGAKEKNLKVKGPVRMPTKTLRITTRKTPCGEG 69
Query 84 TNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
+ T+DRF++RI+KRLIDLHS S++V+QITSI+I+PGVEVEVT+++
Sbjct 70 SKTWDRFQMRIHKRLIDLHSPSEIVKQITSISIEPGVEVEVTIAD 114
> xla:100101334 hypothetical protein LOC100101334
Length=120
Score = 147 bits (370), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 68/113 (60%), Positives = 93/113 (82%), Gaps = 2/113 (1%)
Query 18 EGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAA--KQRKLQTNGPVRLPVKTLRVTT 75
+ L ++Q++HRIRITLTS ++K++E+VC L A + L+ GPVRLP KTL++TT
Sbjct 8 KALAEDDQQIHRIRITLTSLNVKNLEKVCDVLKKRALDHNQHLKVAGPVRLPTKTLKITT 67
Query 76 RKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
RKSPCGEGTNT+DR+++RI+KR+IDLHSASD V+ ITSI I+PGVEVE+T++N
Sbjct 68 RKSPCGEGTNTWDRYQMRIHKRIIDLHSASDAVKHITSIVIEPGVEVEITINN 120
> bbo:BBOV_II004020 18.m06332; 40S ribosomal protein S20; K02969
small subunit ribosomal protein S20e
Length=120
Score = 144 bits (363), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 63/102 (61%), Positives = 85/102 (83%), Gaps = 0/102 (0%)
Query 25 QRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEGT 84
R+H+I +TLTS +LK+IE+ C +I AK++ L+ GPVR+PV+TL++TTRKSP GEGT
Sbjct 19 NRIHKIHVTLTSMNLKAIEKACKTMINGAKEKDLKVVGPVRMPVRTLKITTRKSPVGEGT 78
Query 85 NTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTV 126
NT+D+FE R+YKR+I LHS+S+VV QITS +DPGVEVEVT+
Sbjct 79 NTWDKFEARLYKRVISLHSSSEVVTQITSFPLDPGVEVEVTI 120
> cel:Y105E8A.16 rps-20; Ribosomal Protein, Small subunit family
member (rps-20); K02969 small subunit ribosomal protein S20e
Length=117
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 62/114 (54%), Positives = 90/114 (78%), Gaps = 0/114 (0%)
Query 12 MSKALKEGLEGEEQRLHRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTL 71
M+ A K + HRIR+TLTS+++K +E+VC++LI AK L GP+R+P K L
Sbjct 1 MAVAFKNEKAITDNSEHRIRLTLTSQNVKPLEKVCAQLIDGAKNEHLIVKGPIRMPTKVL 60
Query 72 RVTTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVT 125
R+TTRK+PCGEG+ T+DRF++RI+KRLI+LH+ ++V+RQITSI+I+PGV++EVT
Sbjct 61 RITTRKTPCGEGSKTWDRFQMRIHKRLINLHAPAEVLRQITSISIEPGVDIEVT 114
> sce:YHL015W RPS20, URP2; Rps20p; K02969 small subunit ribosomal
protein S20e
Length=121
Score = 106 bits (265), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 49/98 (50%), Positives = 72/98 (73%), Gaps = 0/98 (0%)
Query 31 RITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEGTNTYDRF 90
RITLTS +K +E V S ++ A+Q L GPVRLP K L+++TRK+P GEG+ T++ +
Sbjct 23 RITLTSTKVKQLENVSSNIVKNAEQHNLVKKGPVRLPTKVLKISTRKTPNGEGSKTWETY 82
Query 91 ELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVTVSN 128
E+RI+KR IDL + +V++IT I I+PGV+VEV V++
Sbjct 83 EMRIHKRYIDLEAPVQIVKRITQITIEPGVDVEVVVAS 120
> ath:AT3G13120 30S ribosomal protein S10, chloroplast, putative;
K02946 small subunit ribosomal protein S10
Length=191
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 52/98 (53%), Gaps = 2/98 (2%)
Query 28 HRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEGTNTY 87
+IRI L S + IE C +++ AA+ +T GPV LP K KSP +
Sbjct 93 QKIRIKLRSYWVPLIEDSCKQILDAARNTNAKTMGPVPLPTKKRIYCVLKSPHVHKDARF 152
Query 88 DRFELRIYKRLID-LHSASDVVRQITSINIDPGVEVEV 124
FE+R ++R+ID L+ + + + +++ GV+VEV
Sbjct 153 -HFEIRTHQRMIDILYPTAQTIDSLMQLDLPAGVDVEV 189
> eco:b3321 rpsJ, ECK3308, JW3283, nusE; 30S ribosomal subunit
protein S10; K02946 small subunit ribosomal protein S10
Length=103
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 57/101 (56%), Gaps = 2/101 (1%)
Query 28 HRIRITLTSKDLKSIERVCSELIGAAKQRKLQTNGPVRLPVKTLRVTTRKSPCGEGTNTY 87
RIRI L + D + I++ +E++ AK+ Q GP+ LP + R T SP +
Sbjct 4 QRIRIRLKAFDHRLIDQATAEIVETAKRTGAQVRGPIPLPTRKERFTVLISPHV-NKDAR 62
Query 88 DRFELRIYKRLIDLHSASD-VVRQITSINIDPGVEVEVTVS 127
D++E+R + RL+D+ ++ V + +++ GV+V++++
Sbjct 63 DQYEIRTHLRLVDIVEPTEKTVDALMRLDLAAGVDVQISLG 103
> sce:YPR194C OPT2; Oligopeptide transporter; member of the OPT
family, with potential orthologs in S. Pombe and C. albicans;
also plays a role in formation of mature vacuoles
Length=877
Score = 34.7 bits (78), Expect = 0.086, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 21/42 (50%), Gaps = 0/42 (0%)
Query 82 EGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVE 123
EG + YD F++R + I HS VR + DP + VE
Sbjct 123 EGIDEYDSFDIRAFASAIKFHSPYQEVRAVVDPEDDPTIPVE 164
> xla:495120 phox2b, phox2, xphox2b; paired-like homeobox 2b
Length=293
Score = 28.9 bits (63), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 94 EKRKQRRIRTTFTSAQLKELERVFAE 119
> dre:544654 phox2b; paired-like homeobox 2b; K09330 paired mesoderm
homeobox protein 2
Length=284
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 94 EKRKQRRIRTTFTSAQLKELERVFAE 119
> xla:402788 phox2a, phox2, xphox2a; paired-like homeobox 2a;
K09330 paired mesoderm homeobox protein 2
Length=281
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 88 EKRKQRRIRTTFTSSQLKELERVFAE 113
> xla:402787 phox2b; paired-like homeobox 2b; K09330 paired mesoderm
homeobox protein 2
Length=293
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 94 EKRKQRRIRTTFTSAQLKELERVFAE 119
> ath:AT5G61210 SNAP33; SNAP33 (SOLUBLE N-ETHYLMALEIMIDE-SENSITIVE
FACTOR ADAPTOR PROTEIN 33); SNAP receptor/ protein binding;
K08509 synaptosomal-associated protein, 29kDa
Length=300
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 13/47 (27%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 77 KSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVE 123
+ P E + Y R E+ K+ L SD++ ++ ++ +D G E+E
Sbjct 221 REPLPESADAYQRVEMEKAKQDDGLSDLSDILGELKNMAVDMGSEIE 267
> dre:404602 phox2a, MGC77364, arix, sou, zgc:77364; paired-like
homeobox 2a; K09330 paired mesoderm homeobox protein 2
Length=277
Score = 28.9 bits (63), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 85 EKRKQRRIRTTFTSSQLKELERVFAE 110
> mmu:11859 Phox2a, Arix, Phox2, Pmx2, Pmx2a; paired-like homeobox
2a; K09330 paired mesoderm homeobox protein 2
Length=280
Score = 28.9 bits (63), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 86 EKRKQRRIRTTFTSAQLKELERVFAE 111
> hsa:401 PHOX2A, ARIX, CFEOM2, FEOM2, MGC52227, NCAM2, PMX2A;
paired-like homeobox 2a; K09330 paired mesoderm homeobox protein
2
Length=284
Score = 28.5 bits (62), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 86 EKRKQRRIRTTFTSAQLKELERVFAE 111
> mmu:18935 Phox2b, Dilp1, NBPhox, Pmx2b; paired-like homeobox
2b; K09330 paired mesoderm homeobox protein 2
Length=314
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 94 EKRKQRRIRTTFTSAQLKELERVFAE 119
> hsa:8929 PHOX2B, NBLST2, NBPhox, PMX2B; paired-like homeobox
2b; K09330 paired mesoderm homeobox protein 2
Length=314
Score = 28.5 bits (62), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 23 EEQRLHRIRITLTSKDLKSIERVCSE 48
E+++ RIR T TS LK +ERV +E
Sbjct 94 EKRKQRRIRTTFTSAQLKELERVFAE 119
> mmu:20115 Rps7, Rps7A, S7; ribosomal protein S7 (EC:3.6.5.3);
K02993 small subunit ribosomal protein S7e
Length=194
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 79 PCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEV 124
P GE D FE I + L++L SD+ Q+ +NI E+EV
Sbjct 11 PNGEKP---DEFESGISQALLELEMNSDLKAQLRELNITAAKEIEV 53
> hsa:6201 RPS7, DBA8; ribosomal protein S7 (EC:3.6.5.3); K02993
small subunit ribosomal protein S7e
Length=194
Score = 28.5 bits (62), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 79 PCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEV 124
P GE D FE I + L++L SD+ Q+ +NI E+EV
Sbjct 11 PNGEKP---DEFESGISQALLELEMNSDLKAQLRELNITAAKEIEV 53
> xla:397774 rps7, rpS8A; ribosomal protein S8 (EC:3.6.5.3); K02993
small subunit ribosomal protein S7e
Length=194
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 79 PCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEV 124
P GE D FE I + L++L SD+ Q+ +NI E+EV
Sbjct 11 PNGEKP---DEFESGISQALLELEMNSDLKAQLRELNITAAKEIEV 53
> xla:414368 rps7, MGC53555, dba8, rpS8B; ribosomal protein S7;
K02993 small subunit ribosomal protein S7e
Length=194
Score = 28.1 bits (61), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 3/46 (6%)
Query 79 PCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEV 124
P GE D FE I + L++L SD+ Q+ +NI E+EV
Sbjct 11 PNGEKP---DEFESGISQALLELEMNSDLKAQLRELNITAAKEIEV 53
> dre:562437 ubfd1, MGC162578, im:6905175, zgc:162578; ubiquitin
family domain containing 1
Length=279
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 2 IHPFSSLAASMSKALKEGLEGEEQRLHRIRITLTSK 37
IH + L +M K + +GL EE+ L I++T +K
Sbjct 84 IHALTGLPPAMQKVMYKGLLPEEKTLREIKVTNGAK 119
> sce:YKR020W VPS51, API3, VPS67, WHI6; Component of the GARP
(Golgi-associated retrograde protein) complex, Vps51p-Vps52p-Vps53p-Vps54p,
which is required for the recycling of proteins
from endosomes to the late Golgi; links the (VFT/GARP) complex
to the SNARE Tlg1p
Length=164
Score = 28.1 bits (61), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 34/64 (53%), Gaps = 1/64 (1%)
Query 66 LPVKTLRVTTRKSPCGEGTNTYDRFELRIYKRLIDLHSASDVVRQITSINIDPGVEVEVT 125
LP K L V G+ T T + + IY+ DL +D++++IT+ N D +++ T
Sbjct 98 LPFKRL-VQIHNKLLGKETETNNSIKNTIYENYYDLIKVNDLLKEITNANEDQINKLKQT 156
Query 126 VSNI 129
V ++
Sbjct 157 VESL 160
Lambda K H
0.317 0.133 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2044474180
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40