bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2818_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
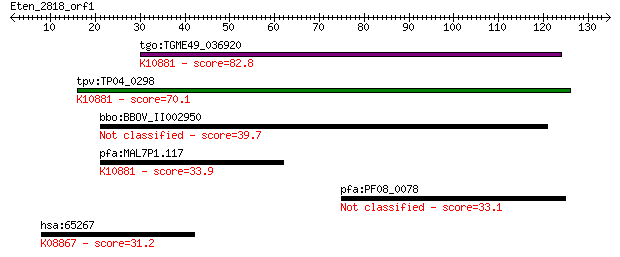
tgo:TGME49_036920 hypothetical protein ; K10881 26 proteasome ... 82.8 3e-16
tpv:TP04_0298 hypothetical protein; K10881 26 proteasome compl... 70.1 2e-12
bbo:BBOV_II002950 18.m06244; 26 proteasome complex subunit SEM1 39.7 0.002
pfa:MAL7P1.117 conserved Plasmodium protein, unknown function;... 33.9 0.13
pfa:PF08_0078 ABC transporter, putative 33.1 0.27
hsa:65267 WNK3, FLJ30437, FLJ42662, KIAA1566, PRKWNK3; WNK lys... 31.2 1.0
> tgo:TGME49_036920 hypothetical protein ; K10881 26 proteasome
complex subunit DSS1
Length=111
Score = 82.8 bits (203), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 48/94 (51%), Positives = 56/94 (59%), Gaps = 25/94 (26%)
Query 30 TGAETQEEAFDEPDDELEEFDEIGVACCTKETMSTQMGDTTERTSESSGGEVPVDAELKQ 89
GAE Q+ FDEPDDELEEFDEIG GG+ VD E+ Q
Sbjct 28 NGAEEQD-TFDEPDDELEEFDEIG------------------------GGDGVVDTEVAQ 62
Query 90 WSEDWDAAGWDDEDPDDEFLEHLQRELEAFKTAY 123
W EDWDAAGWDDED +D+F + LQ+ELEAFK +
Sbjct 63 WDEDWDAAGWDDEDVNDDFCKRLQQELEAFKQRH 96
> tpv:TP04_0298 hypothetical protein; K10881 26 proteasome complex
subunit DSS1
Length=109
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 39/111 (35%), Positives = 58/111 (52%), Gaps = 8/111 (7%)
Query 16 AGEEPQRTDSPAQQTGAETQEEAFDEPDDELEEFDEIGVACCTKETMSTQMGDTTERTSE 75
EE + + T + + FDEPDDELEEFDEIG + + TE
Sbjct 4 TAEETNQVPENKEMTDVSSHFQIFDEPDDELEEFDEIGTILVFLYSFNIIFKYFTENI-- 61
Query 76 SSGGEVPVD-AELKQWSEDWDAAGWDDEDPDDEFLEHLQRELEAFKTAYAS 125
V VD E+ W+EDW+ AGWDDED + +F++ + +ELE ++ + +
Sbjct 62 -----VTVDDPEIADWNEDWETAGWDDEDAESDFVKKILQELENYRKTHNN 107
> bbo:BBOV_II002950 18.m06244; 26 proteasome complex subunit SEM1
Length=85
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 47/100 (47%), Gaps = 24/100 (24%)
Query 21 QRTDSPAQQTGAETQEEAFDEPDDELEEFDEIGVACCTKETMSTQMGDTTERTSESSGGE 80
++T S Q + E F EPD+ELEEFDEI
Sbjct 6 KQTVSVEQDVDMDKDMEVFIEPDNELEEFDEIDTVATVD--------------------- 44
Query 81 VPVDAELKQWSEDWDAAGWDDEDPDDEFLEHLQRELEAFK 120
D E+ W+ DW++AGWDD D D +F++ + +ELE ++
Sbjct 45 ---DPEILNWNSDWESAGWDDVDVDSDFVKRILQELENYR 81
> pfa:MAL7P1.117 conserved Plasmodium protein, unknown function;
K10881 26 proteasome complex subunit DSS1
Length=106
Score = 33.9 bits (76), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 1/41 (2%)
Query 21 QRTDSPAQQTGAETQEEAFDEPDDELEEFDEIGVACCTKET 61
+ D + E FDEPDDELEEF+E+G +T
Sbjct 20 NKNDGKVNENSGENMY-IFDEPDDELEEFEEMGSVALNVDT 59
> pfa:PF08_0078 ABC transporter, putative
Length=1419
Score = 33.1 bits (74), Expect = 0.27, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 75 ESSGGEVPVDAELKQWSEDWDAAGWDDEDPDDEFLEHLQRELEAFKTAYA 124
ES E +++ ++ E+ D G+D+E +D+ EHL+ L K+ Y+
Sbjct 177 ESDQDEYDEESDQDEYDEESDQDGYDEESNEDDLDEHLKERLMKRKSFYS 226
> hsa:65267 WNK3, FLJ30437, FLJ42662, KIAA1566, PRKWNK3; WNK lysine
deficient protein kinase 3 (EC:2.7.11.1); K08867 WNK lysine
deficient protein kinase [EC:2.7.11.1]
Length=1743
Score = 31.2 bits (69), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 22/37 (59%), Gaps = 3/37 (8%)
Query 8 SQMKSQGEAGEEPQRTD---SPAQQTGAETQEEAFDE 41
SQ KS G +PQ T +PAQQTGAE +E D+
Sbjct 509 SQCKSMGNVFPQPQNTTLPLAPAQQTGAECEETEVDQ 545
Lambda K H
0.305 0.122 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2231140792
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40