bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2806_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
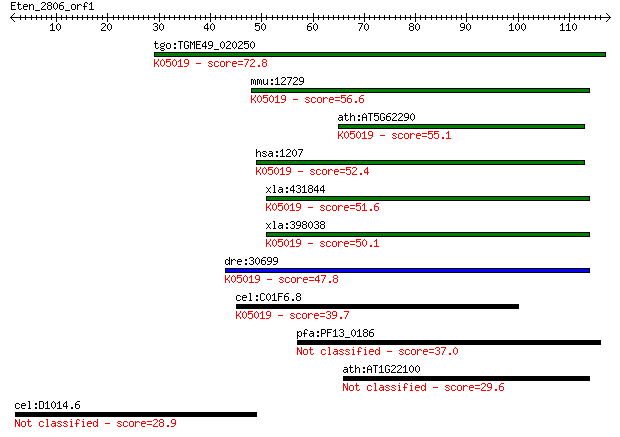
tgo:TGME49_020250 chloride channel, nucleotide-sensitive, 1A, ... 72.8 2e-13
mmu:12729 Clns1a, 2610036D06Rik, 2610100O04Rik, Clci, Clcni, I... 56.6 2e-08
ath:AT5G62290 nucleotide-sensitive chloride conductance regula... 55.1 5e-08
hsa:1207 CLNS1A, CLCI, CLNS1B, ICln; chloride channel, nucleot... 52.4 4e-07
xla:431844 clns1a, MGC81186, clci, clns1b, icln; chloride chan... 51.6 6e-07
xla:398038 clns1a, MGC85110, icln; regulatory protein; K05019 ... 50.1 2e-06
dre:30699 icln; swelling dependent chloride channel; K05019 ch... 47.8 9e-06
cel:C01F6.8 icl-1; ICLn ion channel homolog family member (icl... 39.7 0.002
pfa:PF13_0186 conserved Plasmodium protein, unknown function 37.0 0.016
ath:AT1G22100 ATP binding / inositol pentakisphosphate 2-kinase 29.6 2.4
cel:D1014.6 hypothetical protein 28.9 4.1
> tgo:TGME49_020250 chloride channel, nucleotide-sensitive, 1A,
putative ; K05019 chloride channel, nucleotide-sensitive,
1A
Length=176
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 57/101 (56%), Gaps = 13/101 (12%)
Query 29 TPHRTPSGDLELLH-----NETIGIKVPNTSVVIRGKLAGCGELFVTSQRVAWLA----- 78
+P R G L++L E I + + ++++G+ G G ++TS+R+AWLA
Sbjct 6 SPARDADGSLQILQGPHGDEEVIACREDDAKLILQGENHGVGTFYITSRRIAWLAKPGAA 65
Query 79 ---DNATSFALNYISIVLHALSTDPQACDRPCLYCQIKGDA 116
+ +++Y SIVLHALS DP + PC+YCQ+K DA
Sbjct 66 SGDEQRRDISVDYPSIVLHALSRDPNSGHEPCIYCQLKSDA 106
> mmu:12729 Clns1a, 2610036D06Rik, 2610100O04Rik, Clci, Clcni,
ICLN; chloride channel, nucleotide-sensitive, 1A; K05019 chloride
channel, nucleotide-sensitive, 1A
Length=241
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 38/66 (57%), Gaps = 0/66 (0%)
Query 48 IKVPNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPC 107
++ P+T V+ GK G G L++ R++WL + F+L Y +I LHA+S DP A +
Sbjct 23 LQQPDTEAVLNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHAVSRDPNAYPQEH 82
Query 108 LYCQIK 113
LY +
Sbjct 83 LYVMVN 88
> ath:AT5G62290 nucleotide-sensitive chloride conductance regulator
(ICln) family protein; K05019 chloride channel, nucleotide-sensitive,
1A
Length=228
Score = 55.1 bits (131), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 23/50 (46%), Positives = 37/50 (74%), Gaps = 2/50 (4%)
Query 65 GELFVTSQRVAWLAD--NATSFALNYISIVLHALSTDPQACDRPCLYCQI 112
G L++TS+++ WL+D A +A++++SI LHA+S DP+A PC+Y QI
Sbjct 49 GTLYITSRKLIWLSDVDMAKGYAVDFLSISLHAVSRDPEAYSSPCIYTQI 98
> hsa:1207 CLNS1A, CLCI, CLNS1B, ICln; chloride channel, nucleotide-sensitive,
1A; K05019 chloride channel, nucleotide-sensitive,
1A
Length=237
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 49 KVPNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPCL 108
+ P+T V+ GK G G L++ R++WL + F+L Y +I LHALS D C L
Sbjct 19 QQPDTEAVLNGKGLGTGTLYIAESRLSWLDGSGLGFSLEYPTISLHALSRDRSDCLGEHL 78
Query 109 YCQI 112
Y +
Sbjct 79 YVMV 82
> xla:431844 clns1a, MGC81186, clci, clns1b, icln; chloride channel,
nucleotide-sensitive, 1A; K05019 chloride channel, nucleotide-sensitive,
1A
Length=240
Score = 51.6 bits (122), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 51 PNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPCLYC 110
P T V+ G+ G G L++ R++WL + F+L Y SI LHA+S D A LY
Sbjct 19 PGTEAVVGGRGLGAGTLYIAESRLSWLNSSGLGFSLEYPSISLHAISRDTAAYPEEHLYV 78
Query 111 QIK 113
+
Sbjct 79 MVN 81
> xla:398038 clns1a, MGC85110, icln; regulatory protein; K05019
chloride channel, nucleotide-sensitive, 1A
Length=241
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 33/63 (52%), Gaps = 0/63 (0%)
Query 51 PNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQACDRPCLYC 110
P T V+ G+ G G L++ R++WL + F+L Y SI LHA+S D A LY
Sbjct 19 PGTEAVVGGRGLGPGTLYIAESRLSWLNGSGLGFSLEYPSISLHAISRDTAAYPEEHLYV 78
Query 111 QIK 113
+
Sbjct 79 MVN 81
> dre:30699 icln; swelling dependent chloride channel; K05019
chloride channel, nucleotide-sensitive, 1A
Length=249
Score = 47.8 bits (112), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 36/71 (50%), Gaps = 0/71 (0%)
Query 43 NETIGIKVPNTSVVIRGKLAGCGELFVTSQRVAWLADNATSFALNYISIVLHALSTDPQA 102
+E + ++ T+ V+ GK G G LFV +++W + F L Y +I LHA+S D A
Sbjct 11 SEGVRLQQAETTAVLDGKRLGLGTLFVAEAQLSWFDGSGMGFCLEYPTISLHAISRDLSA 70
Query 103 CDRPCLYCQIK 113
LY +
Sbjct 71 FPEEHLYVMVN 81
> cel:C01F6.8 icl-1; ICLn ion channel homolog family member (icl-1);
K05019 chloride channel, nucleotide-sensitive, 1A
Length=205
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 36/59 (61%), Gaps = 4/59 (6%)
Query 45 TIGIKVPNTSVVIRGKLA--GCGELFVTSQRVAWLADNATS--FALNYISIVLHALSTD 99
T GIK+ T+V K+ G G L++T V W++ A + F++ Y +IVLHA+STD
Sbjct 10 TEGIKLATTNVQAFFKIDSLGNGTLYITDSAVIWISSAAGTKGFSVAYPAIVLHAISTD 68
> pfa:PF13_0186 conserved Plasmodium protein, unknown function
Length=234
Score = 37.0 bits (84), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 41/95 (43%), Gaps = 36/95 (37%)
Query 57 IRGKLA-GCGELFVTSQRVAWLADNA-----TSFA------------------------- 85
I KL G G+L++ +R+ W+ +NA T+F
Sbjct 34 IYNKLNLGEGKLYILEKRLLWINENANKKNVTNFKELCTNNIYLNHYEKNRNFYLHLLNE 93
Query 86 LNYISI-----VLHALSTDPQACDRPCLYCQIKGD 115
+N ISI LHA+++D + CD C+Y Q+ D
Sbjct 94 VNNISIDSSNIALHAITSDKKICDNSCVYIQLNTD 128
> ath:AT1G22100 ATP binding / inositol pentakisphosphate 2-kinase
Length=441
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query 66 ELFVTSQRVAWLADNATSFALNYISIVLH---ALSTDPQACDRPCLYCQIK 113
E +TSQR +W AD A S N S++L L D+PCL +IK
Sbjct 118 EKIITSQRPSWRADVA-SVDTNRSSVLLMDDLTLFAHGHVEDKPCLSVEIK 167
> cel:D1014.6 hypothetical protein
Length=477
Score = 28.9 bits (63), Expect = 4.1, Method: Composition-based stats.
Identities = 21/55 (38%), Positives = 26/55 (47%), Gaps = 9/55 (16%)
Query 2 ILRHSAASIPSIGGFCN--------EELGQRMFEHTPHRTPSGDLELLHNETIGI 48
I+ H + IGGF N E++G M H P T SG L NETIG+
Sbjct 364 IIFHKYQNTSQIGGFWNQPKCIIRPEKIGM-MTIHAPMTTYSGLRRSLVNETIGV 417
Lambda K H
0.322 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2032807080
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40