bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2778_orf1
Length=224
Score E
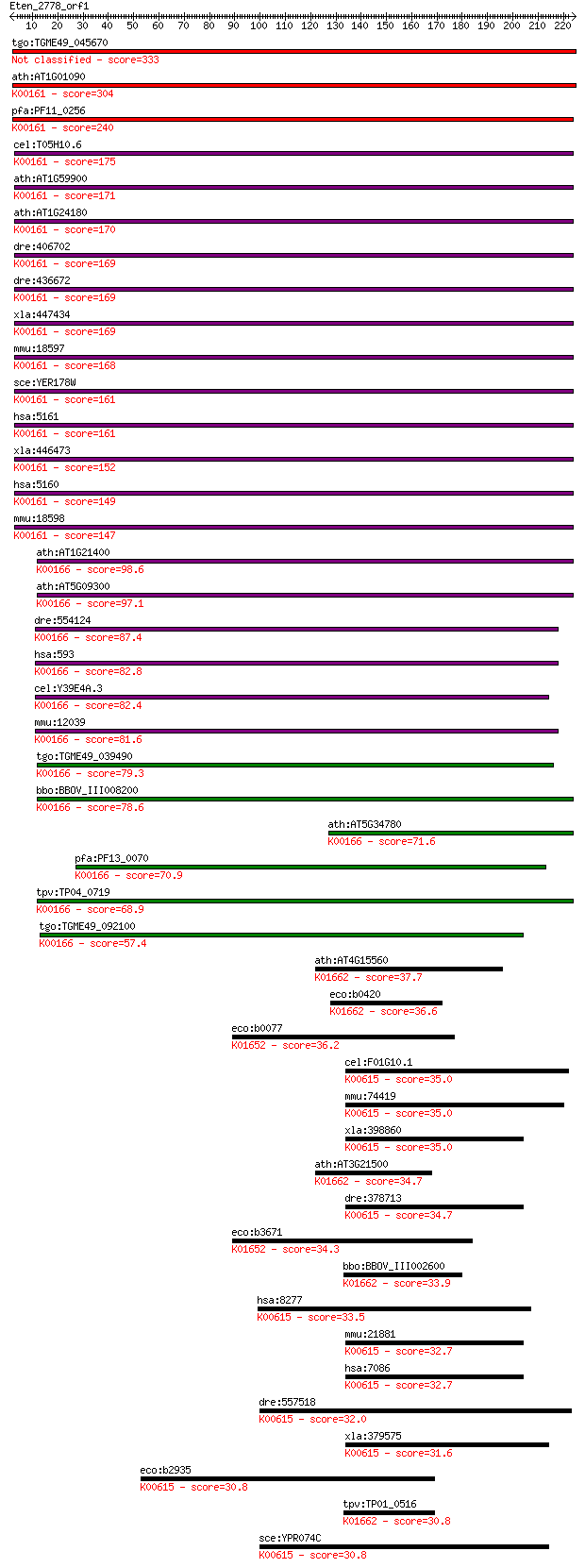
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_045670 pyruvate dehydrogenase, putative (EC:1.2.4.1) 333 3e-91
ath:AT1G01090 PDH-E1 ALPHA (PYRUVATE DEHYDROGENASE E1 ALPHA); ... 304 2e-82
pfa:PF11_0256 pyruvate dehydrogenase E1 component, alpha subun... 240 4e-63
cel:T05H10.6 hypothetical protein; K00161 pyruvate dehydrogena... 175 1e-43
ath:AT1G59900 AT-E1 ALPHA; oxidoreductase, acting on the aldeh... 171 2e-42
ath:AT1G24180 IAR4; IAR4; oxidoreductase, acting on the aldehy... 170 4e-42
dre:406702 pdha1a, im:6895726, pdha1, wu:fd18b01, wu:fo96f03, ... 169 6e-42
dre:436672 pdha1b, Pdha1, wu:fp73b04, zgc:92705; pyruvate dehy... 169 7e-42
xla:447434 pdha1-b, MGC132282, MGC80338, pdha, pdhce1a, phe1a;... 169 8e-42
mmu:18597 Pdha1, Pdha-1; pyruvate dehydrogenase E1 alpha 1 (EC... 168 1e-41
sce:YER178W PDA1; E1 alpha subunit of the pyruvate dehydrogena... 161 1e-39
hsa:5161 PDHA2, MGC149517, MGC149518, PDHAL; pyruvate dehydrog... 161 2e-39
xla:446473 pdha1-a, MGC79036, pdha, pdha1, pdhce1a, phe1a; pyr... 152 1e-36
hsa:5160 PDHA1, PDHA, PDHCE1A, PHE1A; pyruvate dehydrogenase (... 149 9e-36
mmu:18598 Pdha2, Pdhal; pyruvate dehydrogenase E1 alpha 2 (EC:... 147 2e-35
ath:AT1G21400 2-oxoisovalerate dehydrogenase, putative / 3-met... 98.6 2e-20
ath:AT5G09300 2-oxoisovalerate dehydrogenase, putative / 3-met... 97.1 4e-20
dre:554124 bckdha, wu:fd20d04, zgc:110049; branched chain keto... 87.4 3e-17
hsa:593 BCKDHA, BCKDE1A, FLJ45695, MSU, MSUD1, OVD1A; branched... 82.8 9e-16
cel:Y39E4A.3 hypothetical protein; K00166 2-oxoisovalerate deh... 82.4 1e-15
mmu:12039 Bckdha; branched chain ketoacid dehydrogenase E1, al... 81.6 2e-15
tgo:TGME49_039490 mitochondrial branched-chain alpha-keto acid... 79.3 9e-15
bbo:BBOV_III008200 17.m07717; dehydrogenase E1 component famil... 78.6 2e-14
ath:AT5G34780 dehydrogenase E1 component family protein; K0016... 71.6 2e-12
pfa:PF13_0070 branched-chain alpha keto-acid dehydrogenase, pu... 70.9 4e-12
tpv:TP04_0719 branched-chain alpha keto-acid dehydrogenase (EC... 68.9 1e-11
tgo:TGME49_092100 2-oxoisovalerate dehydrogenase, putative (EC... 57.4 4e-08
ath:AT4G15560 CLA1; CLA1 (CLOROPLASTOS ALTERADOS 1); 1-deoxy-D... 37.7 0.036
eco:b0420 dxs, ECK0414, JW0410, yajP; 1-deoxyxylulose-5-phosph... 36.6 0.073
eco:b0077 ilvI, ECK0079, JW0076; acetolactate synthase III, la... 36.2 0.087
cel:F01G10.1 hypothetical protein; K00615 transketolase [EC:2.... 35.0 0.19
mmu:74419 Tktl2, 4933401I19Rik; transketolase-like 2 (EC:2.2.1... 35.0 0.22
xla:398860 tkt, MGC68785; transketolase (EC:2.2.1.1); K00615 t... 35.0 0.23
ath:AT3G21500 DXPS1; 1-deoxy-D-xylulose-5-phosphate synthase (... 34.7 0.26
dre:378713 tkt, cb860, fb38f03, fj52f12, id:ibd3270, wu:cegs27... 34.7 0.31
eco:b3671 ilvB, ECK3662, JW3646; acetolactate synthase I, larg... 34.3 0.32
bbo:BBOV_III002600 17.m07248; 1-deoxy-D-xylulose-5-phosphate s... 33.9 0.49
hsa:8277 TKTL1, TKR, TKT2; transketolase-like 1 (EC:2.2.1.1); ... 33.5 0.70
mmu:21881 Tkt, p68; transketolase (EC:2.2.1.1); K00615 transke... 32.7 1.2
hsa:7086 TKT, FLJ34765, TK, TKT1; transketolase (EC:2.2.1.1); ... 32.7 1.2
dre:557518 Transketolase-like; K00615 transketolase [EC:2.2.1.1] 32.0 1.6
xla:379575 tktl2, MGC69114; transketolase-like 2 (EC:2.2.1.1);... 31.6 2.2
eco:b2935 tktA, ECK2930, JW5478, tkt; transketolase 1, thiamin... 30.8 3.8
tpv:TP01_0516 1-deoxy-D-xylulose 5-phosphate synthase; K01662 ... 30.8 4.4
sce:YPR074C TKL1; Tkl1p (EC:2.2.1.1); K00615 transketolase [EC... 30.8 4.5
> tgo:TGME49_045670 pyruvate dehydrogenase, putative (EC:1.2.4.1)
Length=635
Score = 333 bits (854), Expect = 3e-91, Method: Compositional matrix adjust.
Identities = 173/228 (75%), Positives = 189/228 (82%), Gaps = 9/228 (3%)
Query 2 GRFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVP 61
GR VEDACARLYY GKTAGFVHLYTGQEAVS GV+KLLR DAV STYRDHVHATSKGVP
Sbjct 268 GRMVEDACARLYYMGKTAGFVHLYTGQEAVSAGVIKLLRPDDAVVSTYRDHVHATSKGVP 327
Query 62 AREVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLAR 121
REV AELFGK TGCS+G GGSMHMFSK+ N+ GGFAFIGEQIP+ALG AFS YRR A
Sbjct 328 VREVMAELFGKATGCSRGRGGSMHMFSKKHNMIGGFAFIGEQIPVALGYAFSAAYRRFAM 387
Query 122 RELPGEK-----DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQ 176
G+K DQV VCF+GDGTTNMGQ YEA+N+AAL KLP++FVVENNNWAIGMAAQ
Sbjct 388 ----GDKSDSNADQVAVCFLGDGTTNMGQLYEALNIAALSKLPIVFVVENNNWAIGMAAQ 443
Query 177 RSTAVQEIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGPS 224
RSTA + QR FGV VEVDGMDVLAVR AAR+AI+RAR+GEGP+
Sbjct 444 RSTATPAVWQRAESFGVAGVEVDGMDVLAVRGAARRAIDRARRGEGPT 491
> ath:AT1G01090 PDH-E1 ALPHA (PYRUVATE DEHYDROGENASE E1 ALPHA);
pyruvate dehydrogenase (acetyl-transferring) (EC:1.2.4.1);
K00161 pyruvate dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=428
Score = 304 bits (778), Expect = 2e-82, Method: Compositional matrix adjust.
Identities = 147/223 (65%), Positives = 172/223 (77%), Gaps = 3/223 (1%)
Query 2 GRFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVP 61
GR ED CA++YYRGK GFVHLY GQEAVSTG +KLL K D+V STYRDHVHA SKGV
Sbjct 94 GRSFEDMCAQMYYRGKMFGFVHLYNGQEAVSTGFIKLLTKSDSVVSTYRDHVHALSKGVS 153
Query 62 AREVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLAR 121
AR V +ELFGK TGC +G GGSMHMFSKE N+ GGFAFIGE IP+A G AFS YRR
Sbjct 154 ARAVMSELFGKVTGCCRGQGGSMHMFSKEHNMLGGFAFIGEGIPVATGAAFSSKYRREVL 213
Query 122 RELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAV 181
++ + D VTV F GDGT N GQF+E +NMAAL KLP+IFVVENN WAIGM+ R+T+
Sbjct 214 KQ---DCDDVTVAFFGDGTCNNGQFFECLNMAALYKLPIIFVVENNLWAIGMSHLRATSD 270
Query 182 QEIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGPS 224
EI ++GP FG+P V VDGMDVL VR A++A+ RAR+GEGP+
Sbjct 271 PEIWKKGPAFGMPGVHVDGMDVLKVREVAKEAVTRARRGEGPT 313
> pfa:PF11_0256 pyruvate dehydrogenase E1 component, alpha subunit,
putative; K00161 pyruvate dehydrogenase E1 component subunit
alpha [EC:1.2.4.1]
Length=608
Score = 240 bits (612), Expect = 4e-63, Method: Compositional matrix adjust.
Identities = 118/251 (47%), Positives = 160/251 (63%), Gaps = 29/251 (11%)
Query 2 GRFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVP 61
GR E+ A+LYY + GFVHLY GQEAVSTG++K L+ D V STYRDHVHA SKGVP
Sbjct 203 GRLFENLVAKLYYNKRVNGFVHLYNGQEAVSTGIIKNLKNSDFVTSTYRDHVHALSKGVP 262
Query 62 AREVFAELFGKTTGCS-KGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRR-- 118
A ++ EL+G G + KG GGSMH++SKE N GGF FIGEQIPIA+G+A+S LY+
Sbjct 263 AHKILNELYGNYYGSTNKGKGGSMHIYSKENNFIGGFGFIGEQIPIAVGLAYSILYKNEF 322
Query 119 ------------LARRELPGEKDQ-----------VTVCFMGDGTTNMGQFYEAMNMAAL 155
+ E + V VCF+GDGTTN+GQF+E++N+A+
Sbjct 323 HYNPKNTSFTSTKNKNNYIQENENMIHMNNSQNVDVVVCFLGDGTTNIGQFFESLNLASS 382
Query 156 MKLPVIFVVENNNWAIGMAAQRSTA--VQEIHQRGPPFGVPSVEVDGMDVLAVRAAARQA 213
LP+IFV+ENNNWAIGM + RS++ + + +G F + + +VDG DVL + A++
Sbjct 383 YNLPIIFVIENNNWAIGMESSRSSSDDLMNNYSKGKAFNIDTFKVDGNDVLTIYKLAKKK 442
Query 214 IERAR-KGEGP 223
I++ R + GP
Sbjct 443 IQQIRNRTSGP 453
> cel:T05H10.6 hypothetical protein; K00161 pyruvate dehydrogenase
E1 component subunit alpha [EC:1.2.4.1]
Length=414
Score = 175 bits (444), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 99/221 (44%), Positives = 124/221 (56%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E A LY K GF HLY+GQEA + G+ + +GDAV + YR H G
Sbjct 83 RRMESAAGNLYKEKKIRGFCHLYSGQEACAVGMKAAMTEGDAVITAYRCHGWTWLLGATV 142
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
EV AEL G+ G G GGSMHM++K N YGG +G Q P+ GVA + YR
Sbjct 143 TEVLAELTGRVAGNVHGKGGSMHMYTK--NFYGGNGIVGAQQPLGAGVALAMKYR----- 195
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
E+ V V GDG N GQ +EA NMA L LPV+FV ENN + +G A+RS+A
Sbjct 196 ----EQKNVCVTLYGDGAANQGQLFEATNMAKLWDLPVLFVCENNGFGMGTTAERSSAST 251
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
E + RG VP + VDGMD+LAVR A + A E G+GP
Sbjct 252 EYYTRGDY--VPGIWVDGMDILAVREATKWAKEYCDSGKGP 290
> ath:AT1G59900 AT-E1 ALPHA; oxidoreductase, acting on the aldehyde
or oxo group of donors, disulfide as acceptor / pyruvate
dehydrogenase (acetyl-transferring) (EC:1.2.4.1); K00161
pyruvate dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=389
Score = 171 bits (433), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 92/221 (41%), Positives = 128/221 (57%), Gaps = 12/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E A LY GF HLY GQEAV+ G+ + K DA+ + YRDH +G
Sbjct 70 RRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKDAIITAYRDHCIFLGRGGSL 129
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
EVF+EL G+ GCSKG GGSMH + KE + YGG +G Q+P+ G+AF+Q Y +
Sbjct 130 HEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLGCGIAFAQKYNK---- 185
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
++ VT GDG N GQ +EA+N++AL LP I V ENN++ +G A R+
Sbjct 186 -----EEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEWRAAKSP 240
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
++RG VP ++VDGMD AV+ A + A + A + +GP
Sbjct 241 SYYKRGD--YVPGLKVDGMDAFAVKQACKFAKQHALE-KGP 278
> ath:AT1G24180 IAR4; IAR4; oxidoreductase, acting on the aldehyde
or oxo group of donors, disulfide as acceptor / pyruvate
dehydrogenase (acetyl-transferring) (EC:1.2.4.1); K00161 pyruvate
dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=393
Score = 170 bits (430), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 93/221 (42%), Positives = 126/221 (57%), Gaps = 12/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E A LY GF HLY GQEA++ G+ + K DA+ ++YRDH +G
Sbjct 74 RRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITKKDAIITSYRDHCTFIGRGGKL 133
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+ F+EL G+ TGCS G GGSMH + K+ + YGG +G QIP+ G+AF+Q Y +
Sbjct 134 VDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVGAQIPLGCGLAFAQKYNK---- 189
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
+ VT GDG N GQ +EA+N++AL LP I V ENN++ +G A RS
Sbjct 190 -----DEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTATWRSAKSP 244
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+RG VP ++VDGMD LAV+ A + A E A K GP
Sbjct 245 AYFKRGD--YVPGLKVDGMDALAVKQACKFAKEHALKN-GP 282
> dre:406702 pdha1a, im:6895726, pdha1, wu:fd18b01, wu:fo96f03,
zgc:73271, zgc:86692; pyruvate dehydrogenase (lipoamide) alpha
1a (EC:1.2.4.1); K00161 pyruvate dehydrogenase E1 component
subunit alpha [EC:1.2.4.1]
Length=393
Score = 169 bits (429), Expect = 6e-42, Method: Compositional matrix adjust.
Identities = 89/221 (40%), Positives = 129/221 (58%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HLY GQEA + G+ + D + + YR H + ++G
Sbjct 75 RRMELKADQLYKQKIIRGFCHLYDGQEACAVGIEAGINLSDHLITAYRAHGYTLTRGGTV 134
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
RE+ AEL G+ G +KG GGSMHM++K + YGG +G Q+P+ GVA + Y+
Sbjct 135 REIMAELTGRRGGIAKGKGGSMHMYTKHF--YGGNGIVGAQVPLGAGVALACKYQ----- 187
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
K+++ VC GDG N GQ +E NMA+L KLP IF+ ENN + +G + +R+ A
Sbjct 188 ----GKNELCVCLYGDGAANQGQIFETYNMASLWKLPCIFICENNKYGMGTSVERAAAST 243
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMDVL VR A + A E R G+GP
Sbjct 244 DYYKRGD--FIPGLRVDGMDVLCVREATKFAAEHCRSGKGP 282
> dre:436672 pdha1b, Pdha1, wu:fp73b04, zgc:92705; pyruvate dehydrogenase
(lipoamide) alpha 1b (EC:1.2.4.1); K00161 pyruvate
dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=393
Score = 169 bits (428), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 88/221 (39%), Positives = 131/221 (59%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HLY GQEA + G+ ++ D + + YR H + ++GV
Sbjct 75 RRMELKADQLYKQKIIRGFCHLYDGQEACAVGIEAGIKPTDHLITAYRAHGYTYTRGVSV 134
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+E+ AEL G+ G +KG GGSMHM++K N YGG +G Q+P+ GVA + Y+
Sbjct 135 KEIMAELTGRRGGVAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGVALACQYQ----- 187
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
+++ V GDG N GQ +E+ NMAAL KLP IF+ ENN + +G + +R++A
Sbjct 188 ----GNNEICVTLYGDGAANQGQIFESFNMAALWKLPCIFICENNKYGMGTSVERASAST 243
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMDVL VR A + A + R G+GP
Sbjct 244 DYYKRGD--FIPGLRVDGMDVLGVREATKFAADYCRSGKGP 282
> xla:447434 pdha1-b, MGC132282, MGC80338, pdha, pdhce1a, phe1a;
pyruvate dehydrogenase (lipoamide) alpha 1 (EC:1.2.4.1);
K00161 pyruvate dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=400
Score = 169 bits (427), Expect = 8e-42, Method: Compositional matrix adjust.
Identities = 89/221 (40%), Positives = 128/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HLY GQEA G+ + D + + YR H + ++GV
Sbjct 82 RRMELKSDQLYKQKIIRGFCHLYDGQEACCVGLESGINPTDHLITAYRAHGYTYTRGVSV 141
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+E+ AEL G+ GC+KG GGSMHM++K N YGG +G Q+P+ GVA + +
Sbjct 142 KEILAELTGRRGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGVALACKF------ 193
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
K+++ V GDG N GQ +E NMAAL KLP IF+ ENN + +G + +R+ A
Sbjct 194 ---FGKNEICVSLYGDGAANQGQIFETYNMAALWKLPCIFICENNRYGMGTSVERAAAST 250
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMDVL VR A + A + R G+GP
Sbjct 251 DYYKRGD--YIPGLRVDGMDVLCVREATKFAADHRRSGKGP 289
> mmu:18597 Pdha1, Pdha-1; pyruvate dehydrogenase E1 alpha 1 (EC:1.2.4.1);
K00161 pyruvate dehydrogenase E1 component subunit
alpha [EC:1.2.4.1]
Length=390
Score = 168 bits (426), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 89/221 (40%), Positives = 126/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HL GQEA G+ + D + + YR H ++G+P
Sbjct 72 RRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGINPTDHLITAYRAHGFTFTRGLPV 131
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
R + AEL G+ GC+KG GGSMHM++K N YGG +G Q+P+ G+A + Y
Sbjct 132 RAILAELTGRRGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGIALACKY------ 183
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
KD+V + GDG N GQ +EA NMAAL KLP IF+ ENN + +G + +R+ A
Sbjct 184 ---NGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENNRYGMGTSVERAAAST 240
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMD+L VR A + A R G+GP
Sbjct 241 DYYKRGD--FIPGLRVDGMDILCVREATKFAAAYCRSGKGP 279
> sce:YER178W PDA1; E1 alpha subunit of the pyruvate dehydrogenase
(PDH) complex, catalyzes the direct oxidative decarboxylation
of pyruvate to acetyl-CoA; phosphorylated; regulated
by glucose (EC:1.2.4.1); K00161 pyruvate dehydrogenase E1 component
subunit alpha [EC:1.2.4.1]
Length=420
Score = 161 bits (408), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 86/221 (38%), Positives = 127/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E AC LY K GF HL GQEA++ G+ + K D++ ++YR H +G
Sbjct 92 RRMEMACDALYKAKKIRGFCHLSVGQEAIAVGIENAITKLDSIITSYRCHGFTFMRGASV 151
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+ V AEL G+ G S G GGSMH+++ YGG +G Q+P+ G+AF+ Y+
Sbjct 152 KAVLAELMGRRAGVSYGKGGSMHLYAP--GFYGGNGIVGAQVPLGAGLAFAHQYK----- 204
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
+D + GDG +N GQ +E+ NMA L LPV+F ENN + +G AA RS+A+
Sbjct 205 ----NEDACSFTLYGDGASNQGQVFESFNMAKLWNLPVVFCCENNKYGMGTAASRSSAMT 260
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
E +RG +P ++V+GMD+LAV A++ A + G+GP
Sbjct 261 EYFKRGQ--YIPGLKVNGMDILAVYQASKFAKDWCLSGKGP 299
> hsa:5161 PDHA2, MGC149517, MGC149518, PDHAL; pyruvate dehydrogenase
(lipoamide) alpha 2 (EC:1.2.4.1); K00161 pyruvate dehydrogenase
E1 component subunit alpha [EC:1.2.4.1]
Length=388
Score = 161 bits (407), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 87/221 (39%), Positives = 125/221 (56%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HL GQEA G+ + D V ++YR H ++G+
Sbjct 70 RRMELKADQLYKQKFIRGFCHLCDGQEACCVGLEAGINPSDHVITSYRAHGVCYTRGLSV 129
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
R + AEL G+ GC+KG GGSMHM++K N YGG +G Q P+ G+A + Y+
Sbjct 130 RSILAELTGRRGGCAKGKGGSMHMYTK--NFYGGNGIVGAQGPLGAGIALACKYK----- 182
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
D++ + GDG N GQ EA NMAAL KLP +F+ ENN + +G + +R+ A
Sbjct 183 ----GNDEICLTLYGDGAANQGQIAEAFNMAALWKLPCVFICENNLYGMGTSTERAAASP 238
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P ++VDGMDVL VR A + A R G+GP
Sbjct 239 DYYKRGN--FIPGLKVDGMDVLCVREATKFAANYCRSGKGP 277
> xla:446473 pdha1-a, MGC79036, pdha, pdha1, pdhce1a, phe1a; pyruvate
dehydrogenase (lipoamide) alpha 1 (EC:1.2.4.1); K00161
pyruvate dehydrogenase E1 component subunit alpha [EC:1.2.4.1]
Length=400
Score = 152 bits (383), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 90/221 (40%), Positives = 128/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HLY GQEA G+ + D + + YR H + ++GV
Sbjct 82 RRMELKSDQLYKQKIIRGFCHLYDGQEACCVGLESGINPTDHLITAYRAHGYTYTRGVSV 141
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+E+ AEL G+ GC+KG GGSMHM++K N YGG +G Q+P+ GVA + +
Sbjct 142 KEILAELTGRKGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGVALACKF------ 193
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
KD++ V GDG N GQ +E NMAAL KLP IF+ ENN + +G + +R+ A
Sbjct 194 ---FGKDEICVSLYGDGAANQGQIFETYNMAALWKLPCIFICENNRYGMGTSVERAAAST 250
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMDVL VR A + A + R G+GP
Sbjct 251 DYYKRGD--YIPGLRVDGMDVLCVREATKFAADHCRSGKGP 289
> hsa:5160 PDHA1, PDHA, PDHCE1A, PHE1A; pyruvate dehydrogenase
(lipoamide) alpha 1 (EC:1.2.4.1); K00161 pyruvate dehydrogenase
E1 component subunit alpha [EC:1.2.4.1]
Length=390
Score = 149 bits (375), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 90/221 (40%), Positives = 126/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HL GQEA G+ + D + + YR H ++G+
Sbjct 72 RRMELKADQLYKQKIIRGFCHLCDGQEACCVGLEAGINPTDHLITAYRAHGFTFTRGLSV 131
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
RE+ AEL G+ GC+KG GGSMHM++K N YGG +G Q+P+ G+A + Y
Sbjct 132 REILAELTGRKGGCAKGKGGSMHMYAK--NFYGGNGIVGAQVPLGAGIALACKY------ 183
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
KD+V + GDG N GQ +EA NMAAL KLP IF+ ENN + +G + +R+ A
Sbjct 184 ---NGKDEVCLTLYGDGAANQGQIFEAYNMAALWKLPCIFICENNRYGMGTSVERAAAST 240
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ ++RG +P + VDGMD+L VR A R A R G+GP
Sbjct 241 DYYKRGD--FIPGLRVDGMDILCVREATRFAAAYCRSGKGP 279
> mmu:18598 Pdha2, Pdhal; pyruvate dehydrogenase E1 alpha 2 (EC:1.2.4.1);
K00161 pyruvate dehydrogenase E1 component subunit
alpha [EC:1.2.4.1]
Length=391
Score = 147 bits (372), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 88/221 (39%), Positives = 127/221 (57%), Gaps = 13/221 (5%)
Query 3 RFVEDACARLYYRGKTAGFVHLYTGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPA 62
R +E +LY + GF HL GQEA G+ + D V ++YR H ++G+
Sbjct 73 RRMELKADQLYKQKFIRGFCHLCDGQEACCVGLEAGINPTDHVITSYRAHGFCYTRGLSV 132
Query 63 REVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARR 122
+ + AEL G+ GC+KG GGSMHM+ K N YGG +G Q+P+ GVAF+ Y +
Sbjct 133 KSILAELTGRKGGCAKGKGGSMHMYGK--NFYGGNGIVGAQVPLGAGVAFACKYLK---- 186
Query 123 ELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQ 182
QV + GDG N GQ +EA NM+AL KLP +F+ ENN + +G + +RS A
Sbjct 187 -----NGQVCLALYGDGAANQGQVFEAYNMSALWKLPCVFICENNLYGMGTSNERSAAST 241
Query 183 EIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+ H++G F +P + V+GMD+L VR A + A + R G+GP
Sbjct 242 DYHKKG--FIIPGLRVNGMDILCVREATKFAADHCRSGKGP 280
> ath:AT1G21400 2-oxoisovalerate dehydrogenase, putative / 3-methyl-2-oxobutanoate
dehydrogenase, putative / branched-chain
alpha-keto acid dehydrogenase E1 alpha subunit, putative;
K00166 2-oxoisovalerate dehydrogenase E1 component, alpha subunit
[EC:1.2.4.4]
Length=472
Score = 98.6 bits (244), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 66/215 (30%), Positives = 97/215 (45%), Gaps = 12/215 (5%)
Query 12 LYYRGKTAGFVHLY---TGQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAE 68
++Y + G + Y G+EA++ L D V YR+ +G E +
Sbjct 146 IFYEAQRQGRISFYLTSVGEEAINIASAAALSPDDVVLPQYREPGVLLWRGFTLEEFANQ 205
Query 69 LFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEK 128
FG KG +H S N + + I Q+P A GV +S + K
Sbjct 206 CFGNKADYGKGRQMPIHYGSNRLNYFTISSPIATQLPQAAGVGYSLKMDK---------K 256
Query 129 DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRG 188
+ TV F+GDG T+ G F+ +N AA+M+ PV+F+ NN WAI I +G
Sbjct 257 NACTVTFIGDGGTSEGDFHAGLNFAAVMEAPVVFICRNNGWAISTHISEQFRSDGIVVKG 316
Query 189 PPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+G+ S+ VDG D LAV +A R A E A + P
Sbjct 317 QAYGIRSIRVDGNDALAVYSAVRSAREMAVTEQRP 351
> ath:AT5G09300 2-oxoisovalerate dehydrogenase, putative / 3-methyl-2-oxobutanoate
dehydrogenase, putative / branched-chain
alpha-keto acid dehydrogenase E1 alpha subunit, putative;
K00166 2-oxoisovalerate dehydrogenase E1 component, alpha subunit
[EC:1.2.4.4]
Length=472
Score = 97.1 bits (240), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 63/215 (29%), Positives = 101/215 (46%), Gaps = 12/215 (5%)
Query 12 LYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAE 68
++Y + G + Y G+EA++ L D + YR+ +G +E +
Sbjct 146 IFYEAQRQGRLSFYATAIGEEAINIASAAALTPQDVIFPQYREPGVLLWRGFTLQEFANQ 205
Query 69 LFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEK 128
FG + KG +H S + N + A I Q+P A+G A+S + K
Sbjct 206 CFGNKSDYGKGRQMPVHYGSNKLNYFTVSATIATQLPNAVGAAYSLKMDK---------K 256
Query 129 DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRG 188
D V + GDG T+ G F+ A+N+AA+M+ PV+F+ NN WAI + +G
Sbjct 257 DACAVTYFGDGGTSEGDFHAALNIAAVMEAPVLFICRNNGWAISTPTSDQFRSDGVVVKG 316
Query 189 PPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+G+ S+ VDG D LA+ +A A E A + + P
Sbjct 317 RAYGIRSIRVDGNDALAMYSAVHTAREMAIREQRP 351
> dre:554124 bckdha, wu:fd20d04, zgc:110049; branched chain keto
acid dehydrogenase E1, alpha polypeptide (EC:1.2.4.4); K00166
2-oxoisovalerate dehydrogenase E1 component, alpha subunit
[EC:1.2.4.4]
Length=446
Score = 87.4 bits (215), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 60/210 (28%), Positives = 92/210 (43%), Gaps = 12/210 (5%)
Query 11 RLYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFA 67
R+ Y + G + Y G+E G L D V YR+ +G P A
Sbjct 118 RILYESQRQGRISFYMTNYGEEGTHIGSAAALDPSDLVFGQYREAGVLMYRGFPLDLFMA 177
Query 68 ELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGE 127
+ + KG +H SK+ N + + QIP A G A++ +RE
Sbjct 178 QCYANADDLGKGRQMPVHYGSKDLNFVTISSPLATQIPQAAGAAYA------VKRE---N 228
Query 128 KDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQR 187
++V +C+ G+G + G + N +A ++ P+IF NN +AI I R
Sbjct 229 ANRVVICYFGEGAASEGDAHAGFNFSATLECPLIFFCRNNGYAISTPTNEQYRGDGIAAR 288
Query 188 GPPFGVPSVEVDGMDVLAVRAAARQAIERA 217
GP +G+ S+ VDG DV AV A ++A RA
Sbjct 289 GPGYGLMSIRVDGNDVFAVYNATKEARRRA 318
> hsa:593 BCKDHA, BCKDE1A, FLJ45695, MSU, MSUD1, OVD1A; branched
chain keto acid dehydrogenase E1, alpha polypeptide (EC:1.2.4.4);
K00166 2-oxoisovalerate dehydrogenase E1 component,
alpha subunit [EC:1.2.4.4]
Length=445
Score = 82.8 bits (203), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 59/210 (28%), Positives = 89/210 (42%), Gaps = 12/210 (5%)
Query 11 RLYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFA 67
R+ Y + G + Y G+E G L D V YR+ + P A
Sbjct 117 RILYESQRQGRISFYMTNYGEEGTHVGSAAALDNTDLVFGQYREAGVLMYRDYPLELFMA 176
Query 68 ELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGE 127
+ +G + KG +H KE + + + QIP A+G A++ R
Sbjct 177 QCYGNISDLGKGRQMPVHYGCKERHFVTISSPLATQIPQAVGAAYAAKRANANR------ 230
Query 128 KDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQR 187
V +C+ G+G + G + N AA ++ P+IF NN +AI I R
Sbjct 231 ---VVICYFGEGAASEGDAHAGFNFAATLECPIIFFCRNNGYAISTPTSEQYRGDGIAAR 287
Query 188 GPPFGVPSVEVDGMDVLAVRAAARQAIERA 217
GP +G+ S+ VDG DV AV A ++A RA
Sbjct 288 GPGYGIMSIRVDGNDVFAVYNATKEARRRA 317
> cel:Y39E4A.3 hypothetical protein; K00166 2-oxoisovalerate dehydrogenase
E1 component, alpha subunit [EC:1.2.4.4]
Length=432
Score = 82.4 bits (202), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 55/206 (26%), Positives = 91/206 (44%), Gaps = 11/206 (5%)
Query 11 RLYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFA 67
R+ Y + G + Y G+E G L D + YR+ +G
Sbjct 101 RILYDSQRQGRISFYMTSFGEEGNHVGSAAALEPQDLIYGQYREAGVLLWRGYTMENFMN 160
Query 68 ELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGE 127
+ +G KG MH +KE N + + Q+P A+G A++ ++ +
Sbjct 161 QCYGNADDLGKGRQMPMHFGTKERNFVTISSPLTTQLPQAVGSAYAFKQQK--------D 212
Query 128 KDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQR 187
+++ V + GDG + G + A N AA +K P+IF NN +AI I +
Sbjct 213 NNRIAVVYFGDGAASEGDAHAAFNFAATLKCPIIFFCRNNGYAISTPTSEQYGGDGIAGK 272
Query 188 GPPFGVPSVEVDGMDVLAVRAAARQA 213
GP +G+ ++ VDG D+LAV A ++A
Sbjct 273 GPAYGLHTIRVDGNDLLAVYNATKEA 298
> mmu:12039 Bckdha; branched chain ketoacid dehydrogenase E1,
alpha polypeptide (EC:1.2.4.4); K00166 2-oxoisovalerate dehydrogenase
E1 component, alpha subunit [EC:1.2.4.4]
Length=446
Score = 81.6 bits (200), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 57/210 (27%), Positives = 89/210 (42%), Gaps = 12/210 (5%)
Query 11 RLYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFA 67
R+ Y + G + Y G+E G L + D V YR+ + P +
Sbjct 118 RILYESQRQGRISFYMTNYGEEGTHVGSAAALERTDLVFGQYREAGVLMYRDYPLELFMS 177
Query 68 ELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGE 127
+ +G KG +H KE + + + QIP A+G A++ R
Sbjct 178 QCYGNVNDPGKGRQMPVHYGCKERHFVTISSPLATQIPQAVGAAYAAKRANANR------ 231
Query 128 KDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQR 187
+ +C+ G+G + G + N AA ++ P+IF NN +AI I R
Sbjct 232 ---IVICYFGEGAASEGDAHAGFNFAATLECPIIFFCRNNGYAISTPTSEQYRGDGIAAR 288
Query 188 GPPFGVPSVEVDGMDVLAVRAAARQAIERA 217
GP +G+ S+ VDG DV AV A ++A RA
Sbjct 289 GPGYGIMSIRVDGNDVFAVYNATKEARRRA 318
> tgo:TGME49_039490 mitochondrial branched-chain alpha-keto acid
dehydrogenase E1, putative (EC:1.2.4.4); K00166 2-oxoisovalerate
dehydrogenase E1 component, alpha subunit [EC:1.2.4.4]
Length=463
Score = 79.3 bits (194), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 60/207 (28%), Positives = 95/207 (45%), Gaps = 12/207 (5%)
Query 12 LYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAE 68
++Y + G + Y G+EA+ T V L K D + YR+ G A + +
Sbjct 132 MFYSVQRQGRISFYIQNQGEEALQTAVGLALDKKDHLFCQYRELGVLMLHGFTAEDALEQ 191
Query 69 LFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEK 128
LF + SKG + NL+ + Q+P A G ++ +L G+
Sbjct 192 LFARRGDESKGRQMPISYSKHSVNLHTICTPLTTQVPHAAGAGYA--------FKLAGD- 242
Query 129 DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRG 188
D++ V F G+G + G F+ AMN AA +K +FV NN +AI + A I RG
Sbjct 243 DRIAVAFFGEGAASEGDFHAAMNFAATLKSQTLFVCRNNGYAISTPVKDQYAGDGIAIRG 302
Query 189 PPFGVPSVEVDGMDVLAVRAAARQAIE 215
+G+ ++ VDG D+ A A ++A E
Sbjct 303 ISYGMHTIRVDGNDLFASLLATKKARE 329
> bbo:BBOV_III008200 17.m07717; dehydrogenase E1 component family
protein (EC:1.2.4.1); K00166 2-oxoisovalerate dehydrogenase
E1 component, alpha subunit [EC:1.2.4.4]
Length=447
Score = 78.6 bits (192), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 61/215 (28%), Positives = 92/215 (42%), Gaps = 12/215 (5%)
Query 12 LYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAE 68
L+Y + G + Y G+EA+ G L D + YR+ KG + +
Sbjct 117 LWYNIQRQGRISFYIQNQGEEAMQIGCGLALTPEDHIFGQYRELGVLFCKGFTVDDALNQ 176
Query 69 LFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEK 128
LF KG + KE N++ + Q+P A G ++ +LA+
Sbjct 177 LFANKGDECKGRQMPISYSKKECNIHAICTPLTSQLPHAAGAGYA---LKLAK------A 227
Query 129 DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRG 188
+ V F G+G + G F+ AMNMAA+ + IF NN +AI + I RG
Sbjct 228 NACAVGFFGEGAASEGDFHAAMNMAAVRQSQTIFACRNNGYAISTPVRDQYRGDGIAIRG 287
Query 189 PPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+G+PS+ VDG D+ A A + A E K P
Sbjct 288 VAYGMPSIRVDGNDLFASYIATKHAREHCIKHSTP 322
> ath:AT5G34780 dehydrogenase E1 component family protein; K00166
2-oxoisovalerate dehydrogenase E1 component, alpha subunit
[EC:1.2.4.4]
Length=365
Score = 71.6 bits (174), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 53/97 (54%), Gaps = 0/97 (0%)
Query 127 EKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQ 186
EK+ V F+GDG T+ G F+ +N AA+M+ PV+F+ NN WAI I
Sbjct 25 EKNACAVTFIGDGGTSEGDFHAGLNFAAVMEAPVVFICRNNGWAISTHISEQFRSDGIVV 84
Query 187 RGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
+G +G+ S+ VDG D LAV +A A E A + P
Sbjct 85 KGQAYGIRSIRVDGNDALAVYSAVCSAREMAVTEQRP 121
> pfa:PF13_0070 branched-chain alpha keto-acid dehydrogenase,
putative (EC:1.2.4.2); K00166 2-oxoisovalerate dehydrogenase
E1 component, alpha subunit [EC:1.2.4.4]
Length=429
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 50/186 (26%), Positives = 87/186 (46%), Gaps = 9/186 (4%)
Query 27 GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAELFGKTTGCSKGFGGSMHM 86
G+E + G+ K L D + YR+ S+G ++ +LFG KG +
Sbjct 118 GEEGLQFGMGKALSVDDHLYCQYRETGVLLSRGFTYTDILNQLFGTKYDEGKGRQMCICY 177
Query 87 FSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEKDQVTVCFMGDGTTNMGQF 146
K+ N++ +G Q+ A G ++ +L +K V V + GDG+++ G F
Sbjct 178 TKKDLNIHTITTPLGSQLSHAAGCGYA--------LKLKNQK-AVAVTYCGDGSSSEGDF 228
Query 147 YEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFGVPSVEVDGMDVLAV 206
Y A+N A++ + +FV +NN +AI + + I R G+ S+ VDG D+ A
Sbjct 229 YAALNFASVRQSQTMFVCKNNLYAISTSIKDQYRGDGIAPRALALGIESIRVDGNDLFAS 288
Query 207 RAAARQ 212
A ++
Sbjct 289 YLATKK 294
> tpv:TP04_0719 branched-chain alpha keto-acid dehydrogenase (EC:1.2.4.4);
K00166 2-oxoisovalerate dehydrogenase E1 component,
alpha subunit [EC:1.2.4.4]
Length=464
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 62/225 (27%), Positives = 93/225 (41%), Gaps = 22/225 (9%)
Query 12 LYYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYR----DHVHATS------K 58
L+Y + G + Y G+EA G L+ D + YR D+ + K
Sbjct 122 LFYNIQRQGRISFYIQNQGEEATQLGAGLALQPQDHLFCQYRYFTKDYKNFRELGVIYVK 181
Query 59 GVPAREVFAELFGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRR 118
G +V A+LF KG + KE NL+ + QIP A G ++
Sbjct 182 GCTEDDVLAQLFSTHKDEGKGRQMPISYSKKEVNLHTITTPLSSQIPQASGSGYA----- 236
Query 119 LARRELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRS 178
++ G D V + F G+G + G + AMN AA+ + IF NN+++I +
Sbjct 237 ---LKMQG-ADAVAMVFFGEGAASEGDCHAAMNFAAVRQAQTIFACRNNSYSISTPVRDQ 292
Query 179 TAVQEIHQRGPPFGVPSVEVDGMDVLAVRAAARQAIERARKGEGP 223
I RG G+PS+ VDG D+ A A + E K P
Sbjct 293 YIGDGIAIRGVALGIPSIRVDGNDLFASYMATKYCREYCVKHSTP 337
> tgo:TGME49_092100 2-oxoisovalerate dehydrogenase, putative (EC:1.2.4.4);
K00166 2-oxoisovalerate dehydrogenase E1 component,
alpha subunit [EC:1.2.4.4]
Length=516
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 45/194 (23%), Positives = 79/194 (40%), Gaps = 12/194 (6%)
Query 13 YYRGKTAGFVHLYT---GQEAVSTGVLKLLRKGDAVASTYRDHVHATSKGVPAREVFAEL 69
+Y + G + Y G+EA G L K D + YR+ +G+ ++ A+L
Sbjct 179 FYDIQRQGRISFYMTSFGEEASLVGSAAALHKDDLLLLQYRELSALMWRGLTLDDILAQL 238
Query 70 FGKTTGCSKGFGGSMHMFSKEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEKD 129
F KG +H + N+ + + +IP GV ++ ++ K+
Sbjct 239 FATKNDPGKGRQMPVHYGATNVNMMPICSPLAVKIPQGAGVGYAYTLQK---------KN 289
Query 130 QVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRGP 189
V V + G+G + G N AA + +F+ NN +AI + R
Sbjct 290 AVAVVYFGEGAASEGDASVGFNFAATLGSQTLFLCRNNGYAISTPVGEQYKGDGVGARAV 349
Query 190 PFGVPSVEVDGMDV 203
+G+ +V VDG D+
Sbjct 350 AYGIDTVRVDGTDL 363
> ath:AT4G15560 CLA1; CLA1 (CLOROPLASTOS ALTERADOS 1); 1-deoxy-D-xylulose-5-phosphate
synthase (EC:2.2.1.7); K01662 1-deoxy-D-xylulose-5-phosphate
synthase [EC:2.2.1.7]
Length=717
Score = 37.7 bits (86), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 34/74 (45%), Gaps = 8/74 (10%)
Query 122 RELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAIGMAAQRSTAV 181
R+L G+ + V V +GDG GQ YEAMN A + +I ++ +N Q S
Sbjct 203 RDLKGKNNNV-VAVIGDGAMTAGQAYEAMNNAGYLDSDMIVILNDNK-------QVSLPT 254
Query 182 QEIHQRGPPFGVPS 195
+ PP G S
Sbjct 255 ATLDGPSPPVGALS 268
> eco:b0420 dxs, ECK0414, JW0410, yajP; 1-deoxyxylulose-5-phosphate
synthase, thiamine-requiring, FAD-requiring (EC:2.2.1.7);
K01662 1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7]
Length=620
Score = 36.6 bits (83), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 128 KDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNNWAI 171
K++ TVC +GDG G +EAMN A ++ ++ ++ +N +I
Sbjct 142 KNRRTVCVIGDGAITAGMAFEAMNHAGDIRPDMLVILNDNEMSI 185
> eco:b0077 ilvI, ECK0079, JW0076; acetolactate synthase III,
large subunit (EC:2.2.1.6); K01652 acetolactate synthase I/II/III
large subunit [EC:2.2.1.6]
Length=574
Score = 36.2 bits (82), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 41/88 (46%), Gaps = 16/88 (18%)
Query 89 KEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEKDQVTVCFMGDGTTNMGQFYE 148
+ W GG +G +P ALGV + LP ++ VC GDG+ M +
Sbjct 412 RRWINSGGLGTMGFGLPAALGVKMA----------LP---EETVVCVTGDGSIQMN--IQ 456
Query 149 AMNMAALMKLPVIFVVENNNWAIGMAAQ 176
++ A +LPV+ V NN + +GM Q
Sbjct 457 ELSTALQYELPVLVVNLNNRY-LGMVKQ 483
> cel:F01G10.1 hypothetical protein; K00615 transketolase [EC:2.2.1.1]
Length=618
Score = 35.0 bits (79), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 3/89 (3%)
Query 134 CFMGDGTTNMGQFYEAMNMAALMKLP-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG 192
C +GDG + G +EA A++ KL ++ +V+ N A V+ R FG
Sbjct 147 CLLGDGESAEGSVWEAAAFASIYKLDNLVAIVDVNRLGQSQATSLGHDVETYKARFAAFG 206
Query 193 VPSVEVDGMDVLAVRAAARQAIERARKGE 221
++ V+G +V + AA A R+ KG+
Sbjct 207 FNAIIVNGHNVDELLAAYETA--RSTKGK 233
> mmu:74419 Tktl2, 4933401I19Rik; transketolase-like 2 (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=627
Score = 35.0 bits (79), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 30/89 (33%), Positives = 42/89 (47%), Gaps = 5/89 (5%)
Query 134 CFMGDGTTNMGQFYEAMNMAA---LMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRGPP 190
C MGDG ++ G +EA+ A+ L L IF V + + TAV E +R
Sbjct 153 CLMGDGESSEGSVWEALAFASHYNLDNLVAIFDVNRLGQSGTAPLEHCTAVYE--KRCQA 210
Query 191 FGVPSVEVDGMDVLAVRAAARQAIERARK 219
FG + VDG DV A+ A +A + K
Sbjct 211 FGWNTYVVDGHDVEALCQAFWKAAQVKNK 239
> xla:398860 tkt, MGC68785; transketolase (EC:2.2.1.1); K00615
transketolase [EC:2.2.1.1]
Length=627
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 134 CFMGDGTTNMGQFYEAMNMAALMKLP-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG 192
C +GDG + G +EAM A KL ++ + + N A V+ +R FG
Sbjct 151 CLLGDGEVSEGSVWEAMAFAGFYKLDNLVAIFDVNRLGQSDPAPLQHKVEVYQKRCEAFG 210
Query 193 VPSVEVDGMDV 203
SV VDG V
Sbjct 211 WHSVVVDGHSV 221
> ath:AT3G21500 DXPS1; 1-deoxy-D-xylulose-5-phosphate synthase
(EC:2.2.1.7); K01662 1-deoxy-D-xylulose-5-phosphate synthase
[EC:2.2.1.7]
Length=640
Score = 34.7 bits (78), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 122 RELPGEKDQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENN 167
R+L G + V V +GDG GQ YEAMN A + +I ++ +N
Sbjct 193 RDLKGMNNSV-VSVIGDGAMTAGQAYEAMNNAGYLHSNMIVILNDN 237
> dre:378713 tkt, cb860, fb38f03, fj52f12, id:ibd3270, wu:cegs2794,
wu:fb38f03, wu:fj52f12; transketolase (EC:2.2.1.1); K00615
transketolase [EC:2.2.1.1]
Length=625
Score = 34.7 bits (78), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 33/71 (46%), Gaps = 1/71 (1%)
Query 134 CFMGDGTTNMGQFYEAMNMAALMKLP-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG 192
C +GDG + G +EAM A+ KL ++ +++ N A V +R FG
Sbjct 150 CMLGDGECSEGSVWEAMAFASHYKLDNLVAILDVNRLGQSEPAPLQHNVNVYKERCEAFG 209
Query 193 VPSVEVDGMDV 203
+ VDG DV
Sbjct 210 FNTYVVDGHDV 220
> eco:b3671 ilvB, ECK3662, JW3646; acetolactate synthase I, large
subunit (EC:2.2.1.6); K01652 acetolactate synthase I/II/III
large subunit [EC:2.2.1.6]
Length=562
Score = 34.3 bits (77), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 44/95 (46%), Gaps = 16/95 (16%)
Query 89 KEWNLYGGFAFIGEQIPIALGVAFSQLYRRLARRELPGEKDQVTVCFMGDGTTNMGQFYE 148
++W GG +G +P A+G A + R++ +CF GDG+ M +
Sbjct 408 RQWLTSGGLGTMGFGLPAAIGAALANPDRKV-------------LCFSGDGSLMMN--IQ 452
Query 149 AMNMAALMKLPVIFVVENNNWAIGMAAQRSTAVQE 183
M A+ +L V ++ NN A+G+ Q+ + E
Sbjct 453 EMATASENQLDVKIILMNNE-ALGLVHQQQSLFYE 486
> bbo:BBOV_III002600 17.m07248; 1-deoxy-D-xylulose-5-phosphate
synthase family protein (EC:2.2.1.7); K01662 1-deoxy-D-xylulose-5-phosphate
synthase [EC:2.2.1.7]
Length=686
Score = 33.9 bits (76), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 133 VCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENN---NWAIGMAAQRST 179
VC +GDG+ G EA+N +K P++ + +N + GM A+ T
Sbjct 204 VCVIGDGSLTGGMAMEALNYTCTIKSPLLIIYNDNEQSSLPTGMPAKNGT 253
> hsa:8277 TKTL1, TKR, TKT2; transketolase-like 1 (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=590
Score = 33.5 bits (75), Expect = 0.70, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 53/112 (47%), Gaps = 15/112 (13%)
Query 99 FIGEQIPIALGVAFS-QLYRRLARRELPGEKDQVTVCFMGDGTTNMGQFYEAMNMA---A 154
++G+ + +A G+A++ + + R + R C M DG ++ G +EAM A +
Sbjct 89 WLGQGLGVACGMAYTGKYFDRASYR---------VFCLMSDGESSEGSVWEAMAFASYYS 139
Query 155 LMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFGVPSVEVDGMDVLAV 206
L L IF V + + A+ + + +R FG + VDG DV A+
Sbjct 140 LDNLVAIFDVNRLGHSGALPAEHCINIYQ--RRCEAFGWNTYVVDGRDVEAL 189
> mmu:21881 Tkt, p68; transketolase (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=623
Score = 32.7 bits (73), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/71 (29%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Query 134 CFMGDGTTNMGQFYEAMNMAALMKLP-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG 192
C +GDG + G +EAM A + KL ++ + + N A V +R FG
Sbjct 151 CMLGDGEVSEGSVWEAMAFAGIYKLDNLVAIFDINRLGQSDPAPLQHQVDIYQKRCEAFG 210
Query 193 VPSVEVDGMDV 203
++ VDG V
Sbjct 211 WHTIIVDGHSV 221
> hsa:7086 TKT, FLJ34765, TK, TKT1; transketolase (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=623
Score = 32.7 bits (73), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 20/71 (28%), Positives = 34/71 (47%), Gaps = 1/71 (1%)
Query 134 CFMGDGTTNMGQFYEAMNMAALMKLP-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG 192
C +GDG + G +EAM A++ KL ++ +++ N A + +R FG
Sbjct 151 CLLGDGELSEGSVWEAMAFASIYKLDNLVAILDINRLGQSDPAPLQHQMDIYQKRCEAFG 210
Query 193 VPSVEVDGMDV 203
++ VDG V
Sbjct 211 WHAIIVDGHSV 221
> dre:557518 Transketolase-like; K00615 transketolase [EC:2.2.1.1]
Length=628
Score = 32.0 bits (71), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 33/132 (25%), Positives = 59/132 (44%), Gaps = 18/132 (13%)
Query 100 IGEQIPIALGVAFSQLYRRLARRELPGEKDQVTV-CFMGDGTTNMGQFYEAMNMAALMKL 158
+G+ + +A G+A++ Y +K V C +GDG + G +EAM A+ +L
Sbjct 125 LGQGLGVACGMAYTAKY---------FDKSSYRVYCLLGDGEMSEGAVWEAMAFASYYQL 175
Query 159 P-VIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFGVPSVEVDGMDVLAVRAAARQA---- 213
++ +++ N A V++ +R FG ++ VDG V + A Q
Sbjct 176 DNLMAILDINRLGQSDPAPLQHHVEKYQRRCEAFGWHAIIVDGHSVEELCKAMSQPRHQP 235
Query 214 ---IERARKGEG 222
I + KG+G
Sbjct 236 TAIIAKTIKGKG 247
> xla:379575 tktl2, MGC69114; transketolase-like 2 (EC:2.2.1.1);
K00615 transketolase [EC:2.2.1.1]
Length=625
Score = 31.6 bits (70), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 37/83 (44%), Gaps = 5/83 (6%)
Query 134 CFMGDGTTNMGQFYEAMNMAA---LMKLPVIFVVENNNWAIGMAAQRSTAVQEIHQRGPP 190
C +GDG ++ G +EAM A+ L L IF V + Q T + +R
Sbjct 151 CLLGDGESSEGAVWEAMAFASHYHLDNLVAIFDVNRLGQSEAAPLQHQTDI--YMKRCEA 208
Query 191 FGVPSVEVDGMDVLAVRAAARQA 213
FG + VDG DV + A QA
Sbjct 209 FGWNTYVVDGHDVAELCHAFWQA 231
> eco:b2935 tktA, ECK2930, JW5478, tkt; transketolase 1, thiamin-binding
(EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=663
Score = 30.8 bits (68), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 55/122 (45%), Gaps = 17/122 (13%)
Query 53 VHATSKGVPAREV--FAELFGKTTGCSK-GFGGSMHMFSKEWNLYGGFAFIGEQIPIALG 109
+H T +P E+ F +L KT G + G+ + + +G+ I A+G
Sbjct 75 LHLTGYDLPMEELKNFRQLHSKTPGHPEVGYTAGVETTTGP---------LGQGIANAVG 125
Query 110 VAFSQLYRRLARR-ELPGEK--DQVTVCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVEN 166
+A ++ + LA + PG D T FMGDG G +E ++A +KL + +
Sbjct 126 MAIAE--KTLAAQFNRPGHDIVDHYTYAFMGDGCMMEGISHEVCSLAGTLKLGKLIAFYD 183
Query 167 NN 168
+N
Sbjct 184 DN 185
> tpv:TP01_0516 1-deoxy-D-xylulose 5-phosphate synthase; K01662
1-deoxy-D-xylulose-5-phosphate synthase [EC:2.2.1.7]
Length=758
Score = 30.8 bits (68), Expect = 4.4, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 133 VCFMGDGTTNMGQFYEAMNMAALMKLPVIFVVENNN 168
+ +GDG G YE++N A +K PVI + +N+
Sbjct 196 ISVIGDGGMTGGMAYESLNYAINIKSPVIVIYNDND 231
> sce:YPR074C TKL1; Tkl1p (EC:2.2.1.1); K00615 transketolase [EC:2.2.1.1]
Length=680
Score = 30.8 bits (68), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 53/118 (44%), Gaps = 5/118 (4%)
Query 100 IGEQIPIALGVAFSQLYRRLARRELPG--EKDQVTVCFMGDGTTNMGQFYEAMNMAALMK 157
+G+ I A+G+A +Q A PG D T F+GDG G EA ++A +K
Sbjct 118 LGQGISNAVGMAMAQA-NLAATYNKPGFTLSDNYTYVFLGDGCLQEGISSEASSLAGHLK 176
Query 158 LPVIFVVENNNWAIGMAAQRSTAVQEIHQRGPPFG--VPSVEVDGMDVLAVRAAARQA 213
L + + ++N A + +++ +R +G V VE D+ + A QA
Sbjct 177 LGNLIAIYDDNKITIDGATSISFDEDVAKRYEAYGWEVLYVENGNEDLAGIAKAIAQA 234
Lambda K H
0.321 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7459475120
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40