bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2760_orf1
Length=180
Score E
Sequences producing significant alignments: (Bits) Value
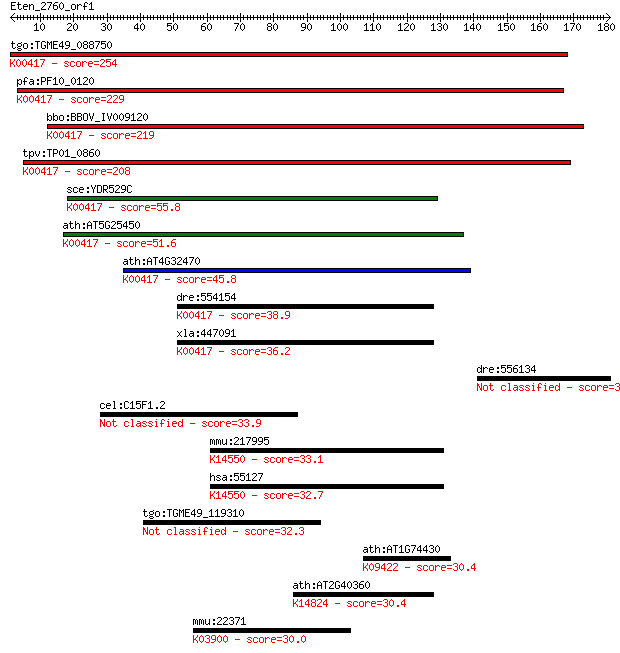
tgo:TGME49_088750 ubiquinol-cytochrome C reductase complex 14 ... 254 1e-67
pfa:PF10_0120 ubiquinol-cytochrome c reductase complex subunit... 229 5e-60
bbo:BBOV_IV009120 23.m06402; hypothetical protein; K00417 ubiq... 219 3e-57
tpv:TP01_0860 ubiquinol-cytochrome C reductase complex 14kD su... 208 9e-54
sce:YDR529C QCR7, COR4, CRO1, UCR7; Qcr7p; K00417 ubiquinol-cy... 55.8 7e-08
ath:AT5G25450 ubiquinol-cytochrome C reductase complex 14 kDa ... 51.6 1e-06
ath:AT4G32470 ubiquinol-cytochrome C reductase complex 14 kDa ... 45.8 9e-05
dre:554154 uqcrb, im:6900881, zgc:109893; ubiquinol-cytochrome... 38.9 0.009
xla:447091 MGC85256 protein; K00417 ubiquinol-cytochrome c red... 36.2 0.068
dre:556134 si:ch211-145c23.4 33.9 0.27
cel:C15F1.2 hypothetical protein 33.9 0.28
mmu:217995 Heatr1, AA517551, B130016L12Rik, BC019693, MGC30806... 33.1 0.48
hsa:55127 HEATR1, BAP28, FLJ10359, MGC72083; HEAT repeat conta... 32.7 0.67
tgo:TGME49_119310 hypothetical protein 32.3 1.0
ath:AT1G74430 MYB95; MYB95 (myb domain protein 95); DNA bindin... 30.4 3.6
ath:AT2G40360 transducin family protein / WD-40 repeat family ... 30.4 3.7
mmu:22371 Vwf, 6820430P06Rik, AI551257, B130011O06Rik, C630030... 30.0 4.1
> tgo:TGME49_088750 ubiquinol-cytochrome C reductase complex 14
kDa protein, putative ; K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=234
Score = 254 bits (649), Expect = 1e-67, Method: Compositional matrix adjust.
Identities = 120/167 (71%), Positives = 134/167 (80%), Gaps = 0/167 (0%)
Query 1 SLVAFTPAAPQPSAVRSFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSD 60
S V + P+ P VR LS L PVWF+FFRGPLDRWN A + KYLR+ GLMYDDLYSD
Sbjct 23 SPVQYVPSPPTKGKVRRALSSALMPVWFKFFRGPLDRWNLAVMAKYLRDHGLMYDDLYSD 82
Query 61 KEPVIERALELLPEDLATARYRRIMRATHLNHVRLYLPPNEQNYDPFIPYLAPYVEEAKF 120
KEPV RALELLP D+ AR+RR+MR T+LNH+RLYLP +EQNYDPFIPY+APYVEEAKF
Sbjct 83 KEPVFARALELLPPDIQAARFRRLMRGTYLNHLRLYLPVHEQNYDPFIPYMAPYVEEAKF 142
Query 121 QLQEEEELLGYHMWERRLYSGGCTGLGDFEPGMHFLVSFPNMYGGGG 167
QLQEEEELLGYHMWE YSGG TG GD EPG HFLV+ PN+YG GG
Sbjct 143 QLQEEEELLGYHMWEGVWYSGGVTGFGDKEPGEHFLVALPNLYGAGG 189
> pfa:PF10_0120 ubiquinol-cytochrome c reductase complex subunit,
putative (EC:1.10.2.2); K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=192
Score = 229 bits (583), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 102/164 (62%), Positives = 130/164 (79%), Gaps = 0/164 (0%)
Query 3 VAFTPAAPQPSAVRSFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKE 62
+ + P + S +R +L+ P +F+F R P +RW A K+LRE GLMYDD+YSDK+
Sbjct 22 LGYEPPKKKRSILRELYHKLIFPYYFKFIRAPYERWQFCATTKFLREHGLMYDDMYSDKD 81
Query 63 PVIERALELLPEDLATARYRRIMRATHLNHVRLYLPPNEQNYDPFIPYLAPYVEEAKFQL 122
PVIERA+ LLP+D+ T RYRR++R TH+N++RL+L P+EQNYDP+IPYLAPY+EEAKFQL
Sbjct 82 PVIERAISLLPKDIQTRRYRRMLRGTHINYLRLFLHPSEQNYDPYIPYLAPYIEEAKFQL 141
Query 123 QEEEELLGYHMWERRLYSGGCTGLGDFEPGMHFLVSFPNMYGGG 166
QEEEELLGYH ++RRLYSGG TG GD EPG+HFLVS PN+YG
Sbjct 142 QEEEELLGYHPYDRRLYSGGTTGFGDLEPGLHFLVSIPNLYGAA 185
> bbo:BBOV_IV009120 23.m06402; hypothetical protein; K00417 ubiquinol-cytochrome
c reductase subunit 7 [EC:1.10.2.2]
Length=197
Score = 219 bits (559), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 99/161 (61%), Positives = 123/161 (76%), Gaps = 0/161 (0%)
Query 12 PSAVRSFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPVIERALEL 71
P + + + P FR F GPL+RW+QA + +YLRE GL+YDDL S++EP++ERALE+
Sbjct 36 PGPLTNIYQTFIGPWVFRMFTGPLERWSQACLTRYLREHGLLYDDLMSEREPIVERALEM 95
Query 72 LPEDLATARYRRIMRATHLNHVRLYLPPNEQNYDPFIPYLAPYVEEAKFQLQEEEELLGY 131
LPEDL TAR+RRI R H++ +R+Y P EQNYDPF+PYLAPY+EEAKFQLQEEEELLGY
Sbjct 96 LPEDLKTARFRRIARGMHISTLRMYPPLEEQNYDPFVPYLAPYIEEAKFQLQEEEELLGY 155
Query 132 HMWERRLYSGGCTGLGDFEPGMHFLVSFPNMYGGGGGAHNK 172
H +RR++ GG TG GD EPGMHFL S PN+YG G G K
Sbjct 156 HPQDRRVFCGGTTGFGDMEPGMHFLTSLPNLYGAGMGNMKK 196
> tpv:TP01_0860 ubiquinol-cytochrome C reductase complex 14kD
subunit (EC:1.10.2.2); K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=197
Score = 208 bits (529), Expect = 9e-54, Method: Compositional matrix adjust.
Identities = 98/164 (59%), Positives = 121/164 (73%), Gaps = 0/164 (0%)
Query 5 FTPAAPQPSAVRSFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPV 64
+T P + + +AP +FR GP +RW+ A + +YLRE GL+YDDL ++EP+
Sbjct 29 YTQIQKGPGFFQKVYRKFIAPWYFRAISGPYERWHHACLTRYLREHGLLYDDLMCEREPI 88
Query 65 IERALELLPEDLATARYRRIMRATHLNHVRLYLPPNEQNYDPFIPYLAPYVEEAKFQLQE 124
IERAL +L DLATAR+RRI R ++ +R+Y P EQNYDPFIPYLAP+VEEAKFQLQE
Sbjct 89 IERALSILTPDLATARFRRIARGMQISMLRIYPPLEEQNYDPFIPYLAPFVEEAKFQLQE 148
Query 125 EEELLGYHMWERRLYSGGCTGLGDFEPGMHFLVSFPNMYGGGGG 168
EEELLGYH +RRL+ GG TG GD EPGMHFLVSFP +YGGG G
Sbjct 149 EEELLGYHPQDRRLFCGGTTGFGDLEPGMHFLVSFPCIYGGGMG 192
> sce:YDR529C QCR7, COR4, CRO1, UCR7; Qcr7p; K00417 ubiquinol-cytochrome
c reductase subunit 7 [EC:1.10.2.2]
Length=127
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 41/112 (36%), Positives = 58/112 (51%), Gaps = 12/112 (10%)
Query 18 FLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLA 77
LS+L PV +F N A K GL +DDL +++ P+++ AL LPED +
Sbjct 20 VLSKLCVPVANQFI-------NLAGYKKL----GLKFDDLIAEENPIMQTALRRLPEDES 68
Query 78 TARYRRIMRATHLNHVRLYLPPNE-QNYDPFIPYLAPYVEEAKFQLQEEEEL 128
AR RI+RA LP NE +PYL PY+ EA+ +E++EL
Sbjct 69 YARAYRIIRAHQTELTHHLLPRNEWIKAQEDVPYLLPYILEAEAAAKEKDEL 120
> ath:AT5G25450 ubiquinol-cytochrome C reductase complex 14 kDa
protein, putative; K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=122
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 61/125 (48%), Gaps = 13/125 (10%)
Query 17 SFLSELLAPVWFRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPV----IERALELL 72
SFL L+ P + L R + ++ LR GL YDDLY +P+ I+ AL L
Sbjct 3 SFLQRLVDPR-----KNFLARMHMKSVSNRLRRYGLRYDDLY---DPLYDLDIKEALNRL 54
Query 73 PEDLATARYRRIMRATHLNHVRLYLPPNEQNYD-PFIPYLAPYVEEAKFQLQEEEELLGY 131
P ++ AR +R+MRA L+ YLP N Q PF YL + K + E E L
Sbjct 55 PREIVDARNQRLMRAMDLSMKHEYLPDNLQAVQTPFRSYLQDMLALVKRERAEREALGAL 114
Query 132 HMWER 136
+++R
Sbjct 115 PLYQR 119
> ath:AT4G32470 ubiquinol-cytochrome C reductase complex 14 kDa
protein, putative; K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=122
Score = 45.8 bits (107), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 53/106 (50%), Gaps = 2/106 (1%)
Query 35 LDRWNQAAIGKYLREQGLMYDDLYSDKEPV-IERALELLPEDLATARYRRIMRATHLNHV 93
L R + AI LR GL YDDLY + I+ A+ LP ++ AR +R+ RA L+
Sbjct 16 LARMHMKAISTRLRRYGLRYDDLYDQYYSMDIKEAMNRLPREVVDARNQRLKRAMDLSMK 75
Query 94 RLYLPPNEQNYD-PFIPYLAPYVEEAKFQLQEEEELLGYHMWERRL 138
YLP + Q PF YL + + + +E E L +++R L
Sbjct 76 HEYLPKDLQAVQTPFRGYLQDMLALVERESKEREALGALPLYQRTL 121
> dre:554154 uqcrb, im:6900881, zgc:109893; ubiquinol-cytochrome
c reductase binding protein (EC:1.10.2.2); K00417 ubiquinol-cytochrome
c reductase subunit 7 [EC:1.10.2.2]
Length=111
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query 51 GLMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHLNHVRLYLPPNE-QNYDPFIP 109
GLM DD + V E A+ LPE L R R+ RA L+ LP ++ ++ +
Sbjct 31 GLMRDDTIDEGSDVKE-AVRRLPEPLYNERVFRLKRAMDLSMKHQILPKDQWTKFEQDVK 89
Query 110 YLAPYVEEAKFQLQEEEE 127
YL PY++E + +E+EE
Sbjct 90 YLEPYLQEVIRERKEKEE 107
> xla:447091 MGC85256 protein; K00417 ubiquinol-cytochrome c reductase
subunit 7 [EC:1.10.2.2]
Length=111
Score = 36.2 bits (82), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 44/79 (55%), Gaps = 4/79 (5%)
Query 51 GLMYDD-LYSDKEPVIERALELLPEDLATARYRRIMRATHLNHVRLYLPPNE-QNYDPFI 108
GLM DD +Y D + ++ A+ LP + R RI RA ++ + +LP + Y+ +
Sbjct 31 GLMRDDTIYEDDD--VKEAIRRLPPRVYDDRLFRIKRALDVSLRQQHLPKQQWTKYEEDV 88
Query 109 PYLAPYVEEAKFQLQEEEE 127
YL PY++E + +E+EE
Sbjct 89 RYLEPYLKEVIRERKEKEE 107
> dre:556134 si:ch211-145c23.4
Length=701
Score = 33.9 bits (76), Expect = 0.27, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Query 141 GGCTGLGDFEPGMHFLVSFP-NMYGGGGGAHNKAHLSSLVP 180
GG LGD P +H +VSFP ++ GA + +HL S++P
Sbjct 649 GGMETLGDVSPHLHGMVSFPSSVLHSVEGAPSNSHLRSVMP 689
> cel:C15F1.2 hypothetical protein
Length=801
Score = 33.9 bits (76), Expect = 0.28, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 7/59 (11%)
Query 28 FRFFRGPLDRWNQAAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMR 86
F+ +G ++R+ Q GK L DD +K +IE+ALE+L E+ Y+++ R
Sbjct 420 FKEVKGLVERFEQRLKGKEL-------DDEQKNKAKLIEKALEMLLENFDDTDYKKVER 471
> mmu:217995 Heatr1, AA517551, B130016L12Rik, BC019693, MGC30806;
HEAT repeat containing 1; K14550 U3 small nucleolar RNA-associated
protein 10
Length=2143
Score = 33.1 bits (74), Expect = 0.48, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 43/79 (54%), Gaps = 9/79 (11%)
Query 61 KEPVIERALELLPEDLATARYRR--------IMRATHLNHVRLYLPP-NEQNYDPFIPYL 111
++ ++ R L+L+P LA ++++ I R T L ++L QN +PFIP L
Sbjct 1633 RKKMVHRFLKLVPVLLAIVQHKKREAEDEQAINRQTALYTLKLLCKNFGAQNREPFIPVL 1692
Query 112 APYVEEAKFQLQEEEELLG 130
+ V+ + + +EE+ +LG
Sbjct 1693 STAVKLIEPEKKEEKNVLG 1711
> hsa:55127 HEATR1, BAP28, FLJ10359, MGC72083; HEAT repeat containing
1; K14550 U3 small nucleolar RNA-associated protein
10
Length=2144
Score = 32.7 bits (73), Expect = 0.67, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 41/79 (51%), Gaps = 9/79 (11%)
Query 61 KEPVIERALELLPEDLATARYRR--------IMRATHLNHVRLYLPP-NEQNYDPFIPYL 111
K+ ++ R L+L+P+ LA + ++ I R T L ++L +N DPF+P L
Sbjct 1634 KKTIVTRFLKLVPDLLAIVQRKKKEGEEEQAINRQTALYTLKLLCKNFGAENPDPFVPVL 1693
Query 112 APYVEEAKFQLQEEEELLG 130
V+ + +EE+ +LG
Sbjct 1694 NTAVKLIAPERKEEKNVLG 1712
> tgo:TGME49_119310 hypothetical protein
Length=3921
Score = 32.3 bits (72), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 20/53 (37%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Query 41 AAIGKYLREQGLMYDDLYSDKEPVIERALELLPEDLATARYRRIMRATHLNHV 93
A YLREQ + +D+ SD RAL+L D +T+ RI+ A H+N
Sbjct 1756 ALTADYLREQRELANDILSDIMKAGSRALKLYHGDTSTSGMARIL-AMHINKT 1807
> ath:AT1G74430 MYB95; MYB95 (myb domain protein 95); DNA binding
/ transcription factor; K09422 myb proto-oncogene protein,
plant
Length=271
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 107 FIPYLAPYVEEAKFQLQEEEELLGYH 132
++ YL P ++ KF QEEEE++ YH
Sbjct 57 WLNYLRPGIKRGKFTPQEEEEIIKYH 82
> ath:AT2G40360 transducin family protein / WD-40 repeat family
protein; K14824 ribosome biogenesis protein ERB1
Length=753
Score = 30.4 bits (67), Expect = 3.7, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 29/45 (64%), Gaps = 3/45 (6%)
Query 86 RATHLNHV---RLYLPPNEQNYDPFIPYLAPYVEEAKFQLQEEEE 127
++ HL ++ +L LP ++++Y+P + Y+ E+A ++L EE+
Sbjct 304 KSKHLTYIPPPKLKLPGHDESYNPSLEYIPTEEEKASYELMFEED 348
> mmu:22371 Vwf, 6820430P06Rik, AI551257, B130011O06Rik, C630030D09,
F8VWF, VWD; Von Willebrand factor homolog; K03900 von
Willebrand factor
Length=2813
Score = 30.0 bits (66), Expect = 4.1, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 28/53 (52%), Gaps = 6/53 (11%)
Query 56 DLYSDKEPVIERALELLP---EDLATARYRRIM---RATHLNHVRLYLPPNEQ 102
+L+S+ E ++ L L P E++ + Y IM R THL H+ Y P N +
Sbjct 2015 ELHSNMEMAVDGRLVLAPYVGENMEVSIYGAIMYEVRFTHLGHILTYTPQNNE 2067
Lambda K H
0.322 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4859948100
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40