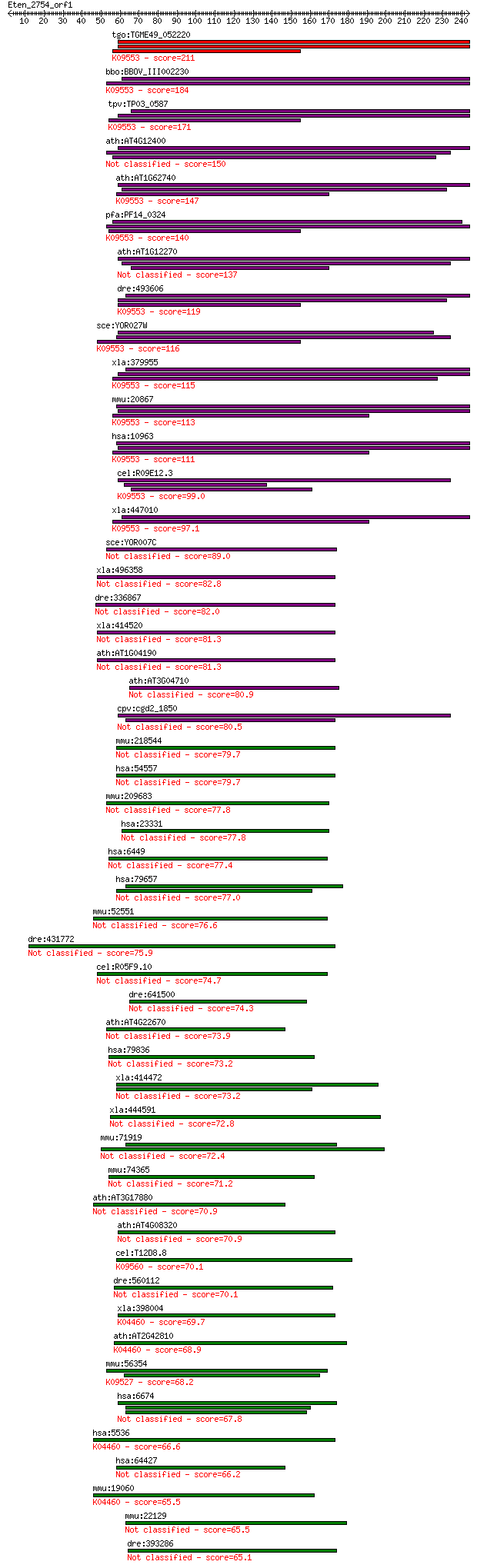
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2754_orf1
Length=243
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_052220 Hsc70/Hsp90-organizing protein, putative ; K... 211 2e-54
bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain ... 184 3e-46
tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phos... 171 2e-42
ath:AT4G12400 stress-inducible protein, putative 150 5e-36
ath:AT1G62740 stress-inducible protein, putative; K09553 stres... 147 3e-35
pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553... 140 3e-33
ath:AT1G12270 stress-inducible protein, putative 137 3e-32
dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1 (... 119 1e-26
sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa gr... 116 7e-26
xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1; K... 115 2e-25
mmu:20867 Stip1, Hop, Sti1, p60; stress-induced phosphoprotein... 113 7e-25
hsa:10963 STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L; stress-i... 111 2e-24
cel:R09E12.3 hypothetical protein; K09553 stress-induced-phosp... 99.0 1e-20
xla:447010 stip1, MGC82554; stress-induced-phosphoprotein 1 (H... 97.1 5e-20
sce:YOR007C SGT2; Glutamine-rich cytoplasmic protein of unknow... 89.0 1e-17
xla:496358 sgta; small glutamine-rich tetratricopeptide repeat... 82.8 1e-15
dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77... 82.0 2e-15
xla:414520 hypothetical protein MGC81394 81.3 3e-15
ath:AT1G04190 tetratricopeptide repeat (TPR)-containing protein 81.3 3e-15
ath:AT3G04710 ankyrin repeat family protein 80.9 4e-15
cpv:cgd2_1850 stress-induced protein sti1-like protein 80.5 5e-15
mmu:218544 Sgtb, C630001O05Rik, MGC27660; small glutamine-rich... 79.7 9e-15
hsa:54557 SGTB, FLJ39002, SGT2; small glutamine-rich tetratric... 79.7 1e-14
mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI85176... 77.8 3e-14
hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28 77.8
hsa:6449 SGTA, SGT, alphaSGT, hSGT; small glutamine-rich tetra... 77.4 4e-14
hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein 3 77.0
mmu:52551 Sgta, 5330427H01Rik, AI194281, D10Ertd190e, MGC6336,... 76.6 8e-14
dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopep... 75.9 1e-13
cel:R05F9.10 sgt-1; Small Glutamine-rich Tetratrico repeat pro... 74.7 3e-13
dre:641500 MGC123010, wu:fk11h08; zgc:123010 74.3 4e-13
ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-inter... 73.9 5e-13
hsa:79836 LONRF3, FLJ22612, MGC119463, MGC119465, RNF127; LON ... 73.2 7e-13
xla:414472 rpap3, MGC81126; RNA polymerase II associated prote... 73.2 8e-13
xla:444591 sgtb, MGC84046; small glutamine-rich tetratricopept... 72.8 1e-12
mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase II... 72.4 2e-12
mmu:74365 Lonrf3, 4932412G04Rik, 5730439E01Rik, A830039N02Rik,... 71.2 3e-12
ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING... 70.9 4e-12
ath:AT4G08320 tetratricopeptide repeat (TPR)-containing protein 70.9 4e-12
cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorig... 70.1 6e-12
dre:560112 si:ch211-243o19.6, wu:fc10f01; si:dkey-33c12.4 70.1 8e-12
xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subuni... 69.7 9e-12
ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphop... 68.9 2e-11
mmu:56354 Dnajc7, 2010003F24Rik, 2010004G07Rik, CCRP, Ttc2, mD... 68.2 3e-11
hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associate... 67.8 3e-11
hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein ph... 66.6 7e-11
hsa:64427 TTC31, FLJ12788, FLJ33201, MGC120200; tetratricopept... 66.2 1e-10
mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalyt... 65.5 2e-10
mmu:22129 Ttc3, 2610202A04Rik, AA409221, D16Ium21, D16Ium21e, ... 65.5 2e-10
dre:393286 ttc25, MGC56362, zgc:56362; tetratricopeptide repea... 65.1 2e-10
> tgo:TGME49_052220 Hsc70/Hsp90-organizing protein, putative ;
K09553 stress-induced-phosphoprotein 1
Length=565
Score = 211 bits (536), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 103/185 (55%), Positives = 134/185 (72%), Gaps = 0/185 (0%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
AQ KAKGNAAFQ G + AV FT AI+ +P D +LYSNRSGA+ASL E AL DAE
Sbjct 7 AQALKAKGNAAFQEGKYEDAVGFFTEAIKCTPDDAVLYSNRSGAYASLNKLEEALNDAEM 66
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
CVKL+P W KGYSRKGLAEF + + EAE +Y KGL+++P+NE L++GL +VQQ
Sbjct 67 CVKLRPTWGKGYSRKGLAEFRMMKYKEAEATYHKGLQVDPTNEQLKEGLNQVQQQTDQFF 126
Query 179 DMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRIREG 238
M A+ A + A+N+HPKL KY Q+DP Y+ L ++L Q+Q NP SL+L+MAQPDVR++EG
Sbjct 127 SMQAMLAAAQAVNKHPKLAKYQQEDPEYTHRLTEILKQIQKNPQSLKLIMAQPDVRVKEG 186
Query 239 MTVLL 243
+ +
Sbjct 187 VIAAM 191
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/190 (30%), Positives = 101/190 (53%), Gaps = 16/190 (8%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A++ + KGN F+ GD+ AA + + AI+ +P+D LYSNR+ A L ++ +AL+DA+T
Sbjct 379 AEQHREKGNEYFKQGDYPAAKKEYDEAIRRNPKDAKLYSNRAAALTKLCEYPSALRDADT 438
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
V++ P + KG+SRKG L +A +++ KGL LEP+N+ +G V
Sbjct 439 SVQVDPAFVKGWSRKGNLHMLLKEYPKALQAFDKGLALEPTNQECIQGKMAV-------- 490
Query 179 DMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQPDV 233
M+ + + ++ P+ + +S DP L +L +Q P + + P
Sbjct 491 -MNKVQQLQSSGEVDPEQMAHSLADPEIQAILKDPQMNIVLMNIQEKPELIHEYLRDP-- 547
Query 234 RIREGMTVLL 243
+I++G+ L+
Sbjct 548 KIKDGINKLI 557
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 51/106 (48%), Gaps = 7/106 (6%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ +A+E K KGN ++ F AA++ + AI+ +P + L +N++ + L D++ L +
Sbjct 241 EQEAEELKQKGNELYKQKKFEAALEAYDEAIEKNPNEILYLNNKAAVYMELGDYDKCLAE 300
Query 116 AETCV----KLKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGL 154
+ + + K D+ AK Y R + G A Y K L
Sbjct 301 CQKALDKRYECKADFSKVAKVYCRMAACKTRSGDYSGAIAMYEKAL 346
> bbo:BBOV_III002230 17.m07215; tetratricopeptide repeat domain
containing protein; K09553 stress-induced-phosphoprotein 1
Length=546
Score = 184 bits (466), Expect = 3e-46, Method: Compositional matrix adjust.
Identities = 93/183 (50%), Positives = 125/183 (68%), Gaps = 1/183 (0%)
Query 61 EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV 120
+ K GN AF+ G F AVQHFT AIQ +P D +LYSNRSGA+ASL+ F+ AL DA CV
Sbjct 3 DHKQLGNEAFKAGRFLDAVQHFTAAIQANPSDGILYSNRSGAYASLQRFQEALDDANQCV 62
Query 121 KLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLRDM 180
LKPDW KGYSRKGLA + LGRL EA +Y +GL+++P+NE L GL+EV +S + M
Sbjct 63 SLKPDWPKGYSRKGLALYKLGRLQEARTAYQEGLKIDPANEPLMSGLREV-ESASDPEFM 121
Query 181 HALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRIREGMT 240
+ A+S + +PKL +Y QQDP+Y L +++S ++ NP SL+ VM P+ IREG+
Sbjct 122 YLSAAMSQLVATNPKLQQYQQQDPSYVMNLCRMISGLKTNPQSLQHVMMDPNPAIREGIM 181
Query 241 VLL 243
+
Sbjct 182 AYI 184
Score = 76.3 bits (186), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 53/196 (27%), Positives = 96/196 (48%), Gaps = 18/196 (9%)
Query 53 DPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENA 112
DP +A+E + KGNA F+ F A + + AI+ +P D LY+NR+ A L ++ +A
Sbjct 356 DP--QKAEEHREKGNAFFKKFQFPEAKKEYDEAIRRNPSDIKLYTNRAAALTKLGEYPSA 413
Query 113 LKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
L D V++ P + K ++RKG L +A +Y KGL L+P+N+ G +
Sbjct 414 LADCNKAVEMDPTFVKAWARKGNLHVLLKEYSKALEAYDKGLALDPNNQECITGKYDC-- 471
Query 173 SGASLRDMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAK-----LLSQVQANPSSLRLV 227
M + A+S + + + + DP + L +L ++ NP+++
Sbjct 472 -------MAKIQAMSQSGTVDEEQYRQAMADPEVQQMLGDPQFQIILKRLSENPAAMNEY 524
Query 228 MAQPDVRIREGMTVLL 243
++ P +I +G+ L+
Sbjct 525 LSDP--KIAKGIQKLM 538
> tpv:TP03_0587 hypothetical protein; K09553 stress-induced-phosphoprotein
1
Length=540
Score = 171 bits (433), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 80/178 (44%), Positives = 118/178 (66%), Gaps = 1/178 (0%)
Query 66 GNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLKPD 125
GN AF+ G F AV+ FT AI+L+P DH+LYSNRSGA+AS+ + AL DA C+ LKPD
Sbjct 8 GNDAFKAGRFMDAVEFFTKAIELNPDDHVLYSNRSGAYASMYMYNEALADANKCIDLKPD 67
Query 126 WAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLRDMHALWA 185
W KGYSRKGL E+ LG +A+ +Y+ GL +P+NE L K L EV+ + + +L
Sbjct 68 WPKGYSRKGLCEYKLGNPEKAKETYNMGLAYDPNNESLNKALLEVENDKSDTY-IQSLLM 126
Query 186 VSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRIREGMTVLL 243
VS + ++PKL KY +QDP Y+ LA+L+S + +P+ L+ ++ P+ +R+G+ +
Sbjct 127 VSQIIQQNPKLRKYQEQDPEYASKLARLVSHMNTDPAVLQQILTDPNPGLRDGLMACM 184
Score = 69.7 bits (169), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 50/194 (25%), Positives = 89/194 (45%), Gaps = 25/194 (12%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A++ + KGN F+ F A + + AI+ +P D LYSNR+ A L ++ +AL D
Sbjct 355 AEQHREKGNEYFKAFKFPEAKKEYDEAIKRNPTDAKLYSNRAAALLKLCEYPSALADCNK 414
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSN----EGLQKGLKEVQQSG 174
++L P + K ++RKG L +A SY KGL+++P+N +G L ++Q+
Sbjct 415 ALELDPTFVKAWARKGNLHVLLKEYHKAMDSYDKGLKVDPNNNECLQGRNNCLNKIQEMN 474
Query 175 ASLRDMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMA 229
D K++ DP + + +L ++ NP ++ +
Sbjct 475 KGEIDEEQ--------------CKHAMADPEVQQIICDPQFQLILKKISENPMTMAEYLK 520
Query 230 QPDVRIREGMTVLL 243
P +I G+ L+
Sbjct 521 DP--KISNGIQKLI 532
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 51/108 (47%), Gaps = 7/108 (6%)
Query 54 PAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENAL 113
P+Q ++ + K +GN ++ F A++ + AI+L P + LL +N++ + + D+E +
Sbjct 215 PSQVESNKYKEEGNNFYKQKKFTEALEMYNKAIELDPNNLLLENNKAAVYLEMGDYEKCI 274
Query 114 KDAETCVKLKPD-------WAKGYSRKGLAEFNLGRLGEAERSYSKGL 154
K + + D +K Y+R + R +A Y K L
Sbjct 275 KTCNDAIDRRYDVMADFTVVSKIYNRLAACYTKMERYDDAILCYQKSL 322
> ath:AT4G12400 stress-inducible protein, putative
Length=558
Score = 150 bits (378), Expect = 5e-36, Method: Compositional matrix adjust.
Identities = 84/193 (43%), Positives = 115/193 (59%), Gaps = 18/193 (9%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A+E K+KGNAAF +GD+ A+ HFT AI LSP +H+LYSNRS ++ASL +E AL DA+
Sbjct 2 AEEAKSKGNAAFSSGDYATAITHFTEAINLSPTNHILYSNRSASYASLHRYEEALSDAKK 61
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
++LKPDW+KGYSR G A L + EA SY KGLE++PSNE L+ GL + +S S +
Sbjct 62 TIELKPDWSKGYSRLGAAFIGLSKFDEAVDSYKKGLEIDPSNEMLKSGLADASRSRVSSK 121
Query 179 DM--------HALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQ 230
+W TA P Y +QD K + ++Q NP++L L M
Sbjct 122 SNPFVDAFQGKEMWEKLTA---DPGTRVYLEQD-----DFVKTMKEIQRNPNNLNLYMK- 172
Query 231 PDVRIREGMTVLL 243
D R+ + + VLL
Sbjct 173 -DKRVMKALGVLL 184
Score = 95.1 bits (235), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/192 (33%), Positives = 96/192 (50%), Gaps = 25/192 (13%)
Query 53 DPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENA 112
DP T A+EE+ KGN F+ + AV+H++ AI+ +P D YSNR+ + L
Sbjct 365 DP--TIAEEEREKGNGFFKEQKYPEAVKHYSEAIKRNPNDVRAYSNRAACYTKLGALPEG 422
Query 113 LKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEV-- 170
LKDAE C++L P + KGYSRKG +F + +A +Y +GL+ +P N+ G++
Sbjct 423 LKDAEKCIELDPSFTKGYSRKGAIQFFMKEYDKAMETYQEGLKHDPKNQEFLDGVRRCVE 482
Query 171 QQSGASLRDMHALWAVSTALNRHPKLLKYSQ----QDPNYSKTLA-----KLLSQVQANP 221
Q + AS D+ P+ LK Q QDP L+ ++L Q NP
Sbjct 483 QINKASRGDL------------TPEELKERQAKAMQDPEVQNILSDPVMRQVLVDFQENP 530
Query 222 SSLRLVMAQPDV 233
+ + M P V
Sbjct 531 KAAQEHMKNPMV 542
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 44/188 (23%), Positives = 89/188 (47%), Gaps = 28/188 (14%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ +A +EK +GN A++ DF AV+H+T A++L +D +NR+ + + +E ++D
Sbjct 227 KEKALKEKGEGNVAYKKKDFGRAVEHYTKAMELDDEDISYLTNRAAVYLEMGKYEECIED 286
Query 116 AETCV----KLKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
+ V +L+ D+ A+ +RKG A + R + + EP+ E QK L
Sbjct 287 CDKAVERGRELRSDFKMIARALTRKGSALVKMARCSK---------DFEPAIETFQKALT 337
Query 169 EVQQSGASLRDMHALWAVSTALNRH----PKLLKYSQQDPN-------YSKTLAKLLSQV 217
E ++ +L+ ++ V L + P + + ++ N Y + + +
Sbjct 338 E-HRNPDTLKKLNDAEKVKKELEQQEYFDPTIAEEEREKGNGFFKEQKYPEAVKHYSEAI 396
Query 218 QANPSSLR 225
+ NP+ +R
Sbjct 397 KRNPNDVR 404
> ath:AT1G62740 stress-inducible protein, putative; K09553 stress-induced-phosphoprotein
1
Length=571
Score = 147 bits (372), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 86/196 (43%), Positives = 119/196 (60%), Gaps = 21/196 (10%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A E KAKGNAAF +GDF++AV HFT+AI L+P +H+L+SNRS A ASL ++ AL DA+
Sbjct 2 ADEAKAKGNAAFSSGDFNSAVNHFTDAINLTPTNHVLFSNRSAAHASLNHYDEALSDAKK 61
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
V+LKPDW KGYSR G A L + EA +YSKGLE++PSNEGL+ GL + + S + R
Sbjct 62 TVELKPDWGKGYSRLGAAHLGLNQFDEAVEAYSKGLEIDPSNEGLKSGLADAKASASRSR 121
Query 179 DMH-----------ALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLV 227
+W+ TA LLK P++ ++ ++Q NPS+L L
Sbjct 122 ASAPNPFGDAFQGPEMWSKLTADPSTRGLLK----QPDF----VNMMKEIQRNPSNLNLY 173
Query 228 MAQPDVRIREGMTVLL 243
+ D R+ + + VLL
Sbjct 174 LQ--DQRVMQALGVLL 187
Score = 100 bits (250), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 65/181 (35%), Positives = 94/181 (51%), Gaps = 21/181 (11%)
Query 61 EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV 120
EE+ KGN F+ + AV+H+T AI+ +P+D YSNR+ + L LKDAE C+
Sbjct 384 EEREKGNDFFKEQKYPDAVRHYTEAIKRNPKDPRAYSNRAACYTKLGAMPEGLKDAEKCI 443
Query 121 KLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE-VQQSGASLRD 179
+L P + KGYSRKG +F + A +Y KGLE +P+N+ L G+K VQQ + R
Sbjct 444 ELDPTFLKGYSRKGAVQFFMKEYDNAMETYQKGLEHDPNNQELLDGVKRCVQQINKANRG 503
Query 180 MHALWAVSTALNRHPKLLKYSQ----QDPNYSKTLA-----KLLSQVQANPSSLRLVMAQ 230
+ P+ LK Q QDP L ++LS +Q NP++ + M
Sbjct 504 -----------DLTPEELKERQAKGMQDPEIQNILTDPVMRQVLSDLQENPAAAQKHMQN 552
Query 231 P 231
P
Sbjct 553 P 553
Score = 60.8 bits (146), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 37/119 (31%), Positives = 64/119 (53%), Gaps = 16/119 (13%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+AQ+EK GNAA++ DF A+QH++ A+++ +D +NR+ + ++ +KD +
Sbjct 242 KAQKEKELGNAAYKKKDFETAIQHYSTAMEIDDEDISYITNRAAVHLEMGKYDECIKDCD 301
Query 118 TCV----KLKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE 169
V +L+ D+ AK +RKG A LG++ + + Y EP + QK L E
Sbjct 302 KAVERGRELRSDYKMVAKALTRKGTA---LGKMAKVSKDY------EPVIQTYQKALTE 351
> pfa:PF14_0324 Hsp70/Hsp90 organizing protein, putative; K09553
stress-induced-phosphoprotein 1
Length=564
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 69/184 (37%), Positives = 111/184 (60%), Gaps = 1/184 (0%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ +AQ K GN FQ G + AV++F++AI P DH+LYSN SGAFASL F AL+
Sbjct 4 KEEAQRLKELGNKCFQEGKYEEAVKYFSDAITNDPLDHVLYSNLSGAFASLGRFYEALES 63
Query 116 AETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGA 175
A C+ +K DW KGY RKG AE L +L AE++Y +GL+++P+N+ LQ L +V+
Sbjct 64 ANKCISIKKDWPKGYIRKGCAEHGLRQLSNAEKTYLEGLKIDPNNKSLQDALSKVRNENM 123
Query 176 SLRDMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRI 235
L + + ++ + P+L Y +++ NY L + + +NP ++R++++ +I
Sbjct 124 -LENAQLIAHLNNIIENDPQLKSYKEENSNYPHELLNTIKSINSNPMNIRIILSTCHPKI 182
Query 236 REGM 239
EG+
Sbjct 183 SEGV 186
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 61/196 (31%), Positives = 101/196 (51%), Gaps = 18/196 (9%)
Query 53 DPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENA 112
DP +A+E K KGN F+N DF A + + AI+ +P D LYSNR+ A L ++ +A
Sbjct 374 DP--DKAEEHKNKGNEYFKNNDFPNAKKEYDEAIRRNPNDAKLYSNRAAALTKLIEYPSA 431
Query 113 LKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
L+D ++L P + K YSRKG F + +A ++Y+KGLEL+P+N+ +G Q+
Sbjct 432 LEDVMKAIELDPTFVKAYSRKGNLHFFMKDYYKALQAYNKGLELDPNNKECLEGY---QR 488
Query 173 SGASLRDMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAK-----LLSQVQANPSSLRLV 227
+ +M V + K S DP + ++ +L ++ NP+S+
Sbjct 489 CAFKIDEMSKSEKVDE------EQFKKSMADPEIQQIISDPQFQIILQKLNENPNSISEY 542
Query 228 MAQPDVRIREGMTVLL 243
+ P +I G+ L+
Sbjct 543 IKDP--KIFNGLQKLI 556
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 56/111 (50%), Gaps = 13/111 (11%)
Query 54 PAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENAL 113
P Q Q E K KGN ++ F A++ + AIQ++P D + + N++ +K+++ A+
Sbjct 238 PEQIQGDEHKLKGNEFYKQKKFDEALKEYEEAIQINPNDIMYHYNKAAVHIEMKNYDKAV 297
Query 114 KDAETCV-------KLKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGL 154
ETC+ K ++ AK Y+R ++ N+ + A +Y K L
Sbjct 298 ---ETCLYAIENRYNFKAEFIQVAKLYNRLAISYINMKKYDLAIEAYRKSL 345
> ath:AT1G12270 stress-inducible protein, putative
Length=572
Score = 137 bits (345), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 81/192 (42%), Positives = 112/192 (58%), Gaps = 14/192 (7%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A+E KAKGNAAF +GDF A+ HFT AI L+P +H+L+SNRS A ASL + AL DA+
Sbjct 2 AEEAKAKGNAAFSSGDFTTAINHFTEAIALAPTNHVLFSNRSAAHASLHQYAEALSDAKE 61
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
+KLKP W KGYSR G A L + A +Y KGL+++P+NE L+ GL + + S A R
Sbjct 62 TIKLKPYWPKGYSRLGAAHLGLNQFELAVTAYKKGLDVDPTNEALKSGLADAEASVARSR 121
Query 179 -------DMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQP 231
D + T L P + QQ P++ ++ ++Q NPSSL L +
Sbjct 122 AAPNPFGDAFQGPEMWTKLTSDPSTRGFLQQ-PDF----VNMMQEIQKNPSSLNLYLK-- 174
Query 232 DVRIREGMTVLL 243
D R+ + + VLL
Sbjct 175 DQRVMQSLGVLL 186
Score = 105 bits (261), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 67/183 (36%), Positives = 95/183 (51%), Gaps = 21/183 (11%)
Query 61 EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV 120
EE+ KGN F+ + A++H+T AI+ +P DH YSNR+ ++ L LKDAE C+
Sbjct 385 EEREKGNDFFKEQKYPEAIKHYTEAIKRNPNDHKAYSNRAASYTKLGAMPEGLKDAEKCI 444
Query 121 KLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE-VQQSGASLRD 179
+L P ++KGYSRK +F L A +Y GLE +PSN+ L G+K VQQ + R
Sbjct 445 ELDPTFSKGYSRKAAVQFFLKEYDNAMETYQAGLEHDPSNQELLDGVKRCVQQINKANRG 504
Query 180 MHALWAVSTALNRHPKLLKYSQ----QDPNYSKTLA-----KLLSQVQANPSSLRLVMAQ 230
+ P+ LK Q QDP L ++LS +Q NPS+ + M
Sbjct 505 -----------DLTPEELKERQAKGMQDPEIQNILTDPVMRQVLSDLQENPSAAQKHMQN 553
Query 231 PDV 233
P V
Sbjct 554 PMV 556
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 57/111 (51%), Gaps = 16/111 (14%)
Query 66 GNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV----K 121
GNAA++ DF A+QH++ AI++ +D +NR+ + + + ++D V +
Sbjct 251 GNAAYKKKDFETAIQHYSTAIEIDDEDISYLTNRAAVYLEMGKYNECIEDCNKAVERGRE 310
Query 122 LKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE 169
L+ D+ A+ +RKG A L ++ + + Y EP+ E QK L E
Sbjct 311 LRSDYKMVARALTRKGTA---LTKMAKCSKDY------EPAIEAFQKALTE 352
> dre:493606 stip1, zgc:92133; stress-induced-phosphoprotein 1
(Hsp70/Hsp90-organizing protein); K09553 stress-induced-phosphoprotein
1
Length=542
Score = 119 bits (298), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 72/186 (38%), Positives = 108/186 (58%), Gaps = 15/186 (8%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K +GN A G+ A++ +T A+ L P +H+L+SNRS A+A D++NALKDA +K+
Sbjct 8 KDQGNKALSAGNLEEAIRCYTEALTLDPSNHVLFSNRSAAYAKKGDYDNALKDACQTIKI 67
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLRDMHA 182
KPDW KGYSRK A LGRL +A+ +Y +GL EPSN+ L++GL+ ++ A + M+
Sbjct 68 KPDWGKGYSRKAAALEFLGRLEDAKATYQEGLRQEPSNQQLKEGLQNMEARLAEKKMMNP 127
Query 183 -----LWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRIRE 237
L+ +R LL DP+Y +LL Q++ PS L + P R+
Sbjct 128 FSIPNLYEKLEGDSRTRALL----SDPSYR----ELLEQLRNKPSELGTKLQDP--RVMT 177
Query 238 GMTVLL 243
++VLL
Sbjct 178 TLSVLL 183
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 60/180 (33%), Positives = 95/180 (52%), Gaps = 21/180 (11%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A EEK KGN AFQ GD+ A++H++ AI+ +P D L+SNR+ + L +F+ ALKD E
Sbjct 359 ALEEKNKGNDAFQKGDYPLAMKHYSEAIKRNPYDAKLFSNRAACYTKLLEFQLALKDCEE 418
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
C+ L + KGY+RKG A + +A Y K LEL+ +++ +GL+
Sbjct 419 CINLDSTFIKGYTRKGAALEAMKDFSKAMDVYQKALELDSNSKEATEGLQRCM------- 471
Query 179 DMHALWAVSTAL-NRHPKLLK-YSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQP 231
VS A+ N P+ +K + DP + ++ +L Q+Q +P +L + P
Sbjct 472 -------VSQAMRNDSPEDVKRRAMADPEVQQIMSDPAMRMILEQMQKDPQALSDHLKNP 524
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/103 (29%), Positives = 53/103 (51%), Gaps = 7/103 (6%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A +EK GNAA++ DF A++H+ AI+ P + SN++ + DF+ + E
Sbjct 224 ALKEKELGNAAYKKKDFATALKHYEEAIKHDPTNMTYLSNQAAVYFEKGDFDKCRELCEK 283
Query 119 CVKL----KPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGL 154
+ + + D+ AK Y+R G + F + EA + ++K L
Sbjct 284 AIDVGRENREDYRQIAKAYARIGNSYFKQEKYKEAVQFFNKSL 326
> sce:YOR027W STI1; Hsp90 cochaperone, interacts with the Ssa
group of the cytosolic Hsp70 chaperones; activates the ATPase
activity of Ssa1p; homolog of mammalian Hop protein; K09553
stress-induced-phosphoprotein 1
Length=589
Score = 116 bits (290), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 70/173 (40%), Positives = 103/173 (59%), Gaps = 10/173 (5%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQ-DHLLYSNRSGAFASLKDFENALKDAE 117
A E K +GNAAF D+ A++ FT AI++S +H+LYSNRS + SLK F +AL DA
Sbjct 5 ADEYKQQGNAAFTAKDYDKAIELFTKAIEVSETPNHVLYSNRSACYTSLKKFSDALNDAN 64
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASL 177
CVK+ P W+KGY+R G A LG L EAE +Y K LEL+ SN+ ++GL +V ++ +
Sbjct 65 ECVKINPSWSKGYNRLGAAHLGLGDLDEAESNYKKALELDASNKAAKEGLDQVHRTQQAR 124
Query 178 RDMHALWAVSTALNRHPKLLKYSQQDPNYSKTL------AKLLSQVQANPSSL 224
+ L T L P L++ +++P S+ + AKL+ Q NP ++
Sbjct 125 QAQPDLGL--TQLFADPNLIENLKKNPKTSEMMKDPQLVAKLIGYKQ-NPQAI 174
Score = 60.5 bits (145), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 48/189 (25%), Positives = 89/189 (47%), Gaps = 24/189 (12%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+A+E + +G F D+ AV+ +T I+ +P+D YSNR+ A A L F A+ D
Sbjct 395 KAEEARLEGKEYFTKSDWPNAVKAYTEMIKRAPEDARGYSNRAAALAKLMSFPEAIADCN 454
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASL 177
++ P++ + Y RK A+ A + Y+ LE + + + EV +G+S
Sbjct 455 KAIEKDPNFVRAYIRKATAQI-------AVKEYASALE---TLDAARTKDAEV-NNGSSA 503
Query 178 RDMHALWAVSTALNRHP--------KLLKYSQQDPNYSK-----TLAKLLSQVQANPSSL 224
R++ L+ ++ P + + + +DP + + +L Q Q NP++L
Sbjct 504 REIDQLYYKASQQRFQPGTSNETPEETYQRAMKDPEVAAIMQDPVMQSILQQAQQNPAAL 563
Query 225 RLVMAQPDV 233
+ M P+V
Sbjct 564 QEHMKNPEV 572
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 58/114 (50%), Gaps = 8/114 (7%)
Query 48 MATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLK 107
M D ++ +A +EKA+GN ++ F A++H+ A +L +D +NR+ A
Sbjct 251 MEVDEDDSKIEADKEKAEGNKFYKARQFDEAIEHYNKAWELH-KDITYLNNRAAAEYEKG 309
Query 108 DFENA---LKDA-ETCVKLKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGL 154
++E A L DA E +++ D+ +K ++R G A LG L + Y K L
Sbjct 310 EYETAISTLNDAVEQGREMRADYKVISKSFARIGNAYHKLGDLKKTIEYYQKSL 363
> xla:379955 stip1, MGC53256; stress-induced-phosphoprotein 1;
K09553 stress-induced-phosphoprotein 1
Length=543
Score = 115 bits (287), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 71/188 (37%), Positives = 103/188 (54%), Gaps = 19/188 (10%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K KGN A G+ AV+ +T AI+L P++H+LYSNRS A+A K+F AL+D V+L
Sbjct 8 KEKGNKALSAGNLDEAVKCYTEAIKLDPKNHVLYSNRSAAYAKKKEFTKALEDGSKTVEL 67
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLRDMHA 182
K DW KGYSRK A L R EA+++Y +GL EP+N L++G L++M A
Sbjct 68 KADWGKGYSRKAAALEFLNRFEEAKKTYEEGLRHEPTNAQLKEG----------LQNMEA 117
Query 183 LWAVSTALN--RHPKLLKYSQQDPNYSKTLA-----KLLSQVQANPSSLRLVMAQPDVRI 235
A +N P L + + DP L+ +L+ Q++ PS L + P R+
Sbjct 118 RLAEKKFMNPFNSPNLFQKLESDPRTRALLSDPSYKELIEQLRNKPSDLGTKLQDP--RV 175
Query 236 REGMTVLL 243
++VLL
Sbjct 176 MTTLSVLL 183
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 58/193 (30%), Positives = 101/193 (52%), Gaps = 21/193 (10%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A E K KGN +FQ GD+ A++H++ AI+ +P D LYSNR+ + L +F A+KD E
Sbjct 360 ALEAKNKGNESFQKGDYPQAMKHYSEAIKRNPNDAKLYSNRAACYTKLLEFLLAVKDCEE 419
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
C++L+P + KGY+RK A + +A +Y K +EL+ +++ G + S +
Sbjct 420 CIRLEPSFIKGYTRKAAALEAMKDFTKAMDAYQKAMELDSTSKEATDGYQRCMMSQYNRN 479
Query 179 DMHALWAVSTALNRHPKLLK-YSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQPD 232
D +P+ +K + DP + ++ +L Q+Q +P +L + P
Sbjct 480 D-------------NPEDVKRRAMADPEVQQIMSDPAMRLILEQMQKDPQALSDHLKNPV 526
Query 233 V--RIREGMTVLL 243
+ +I++ M V L
Sbjct 527 IAQKIQKLMDVGL 539
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 43/179 (24%), Positives = 85/179 (47%), Gaps = 12/179 (6%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ QAQ+EK GN A++ DF A++H+ A +L P + +N++ + + D+ +
Sbjct 222 KKQAQKEKELGNEAYKKKDFETALKHYGQARELDPANMTYITNQAAVYFEMGDYSKCREL 281
Query 116 AETCVKL----KPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
E +++ + D+ AK Y+R G + F + EA + ++K L + E L+K
Sbjct 282 CEKAIEVGRENREDYRLIAKAYARIGNSYFKEEKNKEAIQFFNKSLAEHRTPEVLKK--- 338
Query 169 EVQQSGASLRDMHALWAVSTALNRHPKLL-KYSQQDPNYSKTLAKLLSQVQANPSSLRL 226
QQ+ L++ + ++ L K S Q +Y + + ++ NP+ +L
Sbjct 339 -CQQAEKILKEQERVAYINPDLALEAKNKGNESFQKGDYPQAMKHYSEAIKRNPNDAKL 396
> mmu:20867 Stip1, Hop, Sti1, p60; stress-induced phosphoprotein
1; K09553 stress-induced-phosphoprotein 1
Length=543
Score = 113 bits (282), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 72/188 (38%), Positives = 105/188 (55%), Gaps = 9/188 (4%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
Q E K KGN A G+ A+Q ++ AI+L PQ+H+LYSNRS A+A D++ A +D
Sbjct 3 QVNELKEKGNKALSAGNIDDALQCYSEAIKLDPQNHVLYSNRSAAYAKKGDYQKAYEDGC 62
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASL 177
V LKPDW KGYSRK A L R EA+R+Y +GL+ E +N L++GL+ ++ A
Sbjct 63 KTVDLKPDWGKGYSRKAAALEFLNRFEEAKRTYEEGLKHEANNLQLKEGLQNMEARLAER 122
Query 178 RDMHA--LWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDVRI 235
+ M+ L + L P+ + DP Y +L+ Q+Q PS L + P R+
Sbjct 123 KFMNPFNLPNLYQKLENDPR-TRSLLSDPTYR----ELIEQLQNKPSDLGTKLQDP--RV 175
Query 236 REGMTVLL 243
++VLL
Sbjct 176 MTTLSVLL 183
Score = 93.2 bits (230), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 104/195 (53%), Gaps = 25/195 (12%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A EEK KGN FQ GD+ A++H+T AI+ +P+D LYSNR+ + L +F+ ALKD E
Sbjct 360 ALEEKNKGNECFQKGDYPQAMKHYTEAIKRNPRDAKLYSNRAACYTKLLEFQLALKDCEE 419
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
C++L+P + KGY+RK A + +A Y K L+L+ S + G + R
Sbjct 420 CIQLEPTFIKGYTRKAAALEAMKDYTKAMDVYQKALDLDSSCKEAADGYQ---------R 470
Query 179 DMHALWAVSTALNRH--PKLLK-YSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQ 230
M A + NRH P+ +K + DP + ++ +L Q+Q +P +L +
Sbjct 471 CMMAQY------NRHDSPEDVKRRAMADPEVQQIMSDPAMRLILEQMQKDPQALSEHLKN 524
Query 231 PDV--RIREGMTVLL 243
P + +I++ M V L
Sbjct 525 PVIAQKIQKLMDVGL 539
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 66/142 (46%), Gaps = 11/142 (7%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ QA +EK GN A++ DF A++H+ A +L P + +N++ D+ +
Sbjct 222 KKQALKEKELGNDAYKKKDFDKALKHYDRAKELDPTNMTYITNQAAVHFEKGDYNKCREL 281
Query 116 AETCVKL----KPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
E +++ + D+ AK Y+R G + F + +A Y+K L + LK
Sbjct 282 CEKAIEVGRENREDYRQIAKAYARIGNSYFKEEKYKDAIHFYNKSL----AEHRTPDVLK 337
Query 169 EVQQSGASLRDMHALWAVSTAL 190
+ QQ+ L++ L ++ L
Sbjct 338 KCQQAEKILKEQERLAYINPDL 359
> hsa:10963 STIP1, HOP, IEF-SSP-3521, P60, STI1, STI1L; stress-induced-phosphoprotein
1; K09553 stress-induced-phosphoprotein
1
Length=543
Score = 111 bits (278), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 70/191 (36%), Positives = 102/191 (53%), Gaps = 15/191 (7%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
Q E K KGN A G+ A+Q ++ AI+L P +H+LYSNRS A+A D++ A +D
Sbjct 3 QVNELKEKGNKALSVGNIDDALQCYSEAIKLDPHNHVLYSNRSAAYAKKGDYQKAYEDGC 62
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASL 177
V LKPDW KGYSRK A L R EA+R+Y +GL+ E +N L++GL+ ++ A
Sbjct 63 KTVDLKPDWGKGYSRKAAALEFLNRFEEAKRTYEEGLKHEANNPQLKEGLQNMEARLAER 122
Query 178 RDMHALWAVSTALNRHPKLLKYSQQDPNY-----SKTLAKLLSQVQANPSSLRLVMAQPD 232
+ M+ P L + + DP T +L+ Q++ PS L + P
Sbjct 123 KFMNPF--------NMPNLYQKLESDPRTRTLLSDPTYRELIEQLRNKPSDLGTKLQDP- 173
Query 233 VRIREGMTVLL 243
RI ++VLL
Sbjct 174 -RIMTTLSVLL 183
Score = 93.6 bits (231), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 104/195 (53%), Gaps = 25/195 (12%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A EEK KGN FQ GD+ A++H+T AI+ +P+D LYSNR+ + L +F+ ALKD E
Sbjct 360 ALEEKNKGNECFQKGDYPQAMKHYTEAIKRNPKDAKLYSNRAACYTKLLEFQLALKDCEE 419
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
C++L+P + KGY+RK A + +A Y K L+L+ S + G + R
Sbjct 420 CIQLEPTFIKGYTRKAAALEAMKDYTKAMDVYQKALDLDSSCKEAADGYQ---------R 470
Query 179 DMHALWAVSTALNRH--PKLLK-YSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQ 230
M A + NRH P+ +K + DP + ++ +L Q+Q +P +L +
Sbjct 471 CMMAQY------NRHDSPEDVKRRAMADPEVQQIMSDPAMRLILEQMQKDPQALSEHLKN 524
Query 231 PDV--RIREGMTVLL 243
P + +I++ M V L
Sbjct 525 PVIAQKIQKLMDVGL 539
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 67/142 (47%), Gaps = 11/142 (7%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ QA +EK GN A++ DF A++H+ A +L P + +N++ + D+ +
Sbjct 222 KKQALKEKELGNDAYKKKDFDTALKHYDKAKELDPTNMTYITNQAAVYFEKGDYNKCREL 281
Query 116 AETCVKL----KPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
E +++ + D+ AK Y+R G + F + +A Y+K L + LK
Sbjct 282 CEKAIEVGRENREDYRQIAKAYARIGNSYFKEEKYKDAIHFYNKSL----AEHRTPDVLK 337
Query 169 EVQQSGASLRDMHALWAVSTAL 190
+ QQ+ L++ L ++ L
Sbjct 338 KCQQAEKILKEQERLAYINPDL 359
> cel:R09E12.3 hypothetical protein; K09553 stress-induced-phosphoprotein
1
Length=320
Score = 99.0 bits (245), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 54/175 (30%), Positives = 95/175 (54%), Gaps = 10/175 (5%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
AQEEK KGN F+ GD+ A++H+ A++ P++ +LYSNR+ L +F+ AL D +T
Sbjct 140 AQEEKNKGNEYFKKGDYPTAMRHYNEAVKRDPENAILYSNRAACLTKLMEFQRALDDCDT 199
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
C++L + KGY RK + +A+R+Y L+++PSNE ++G++ +S
Sbjct 200 CIRLDSKFIKGYIRKAACLVAMREWSKAQRAYEDALQVDPSNEEAREGVRNCLRSNDE-- 257
Query 179 DMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAKLLSQVQANPSSLRLVMAQPDV 233
D S A ++L+ DP + +L Q+ +P ++R + P++
Sbjct 258 DPEKAKERSLADPEVQEILR----DPG----MRMILEQMSNDPGAVREHLKNPEI 304
Score = 43.9 bits (102), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 7/82 (8%)
Query 62 EKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVK 121
EK GNAA++ DF A H+ AI+L P + Y+N++ + K F ++ E V+
Sbjct 8 EKDLGNAAYKQKDFEKAHVHYDKAIELDPSNITFYNNKAAVYFEEKKFAECVQFCEKAVE 67
Query 122 L----KPDW---AKGYSRKGLA 136
+ + D+ AK SR G A
Sbjct 68 VGRETRADYKLIAKAMSRAGNA 89
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/95 (27%), Positives = 40/95 (42%), Gaps = 8/95 (8%)
Query 66 GNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLKPD 125
GNA + D AVQ F ++ L+ +K+ E LK AE + P+
Sbjct 87 GNAFQKQNDLSLAVQWFHRSLSEFRDPELV--------KKVKELEKQLKAAERLAYINPE 138
Query 126 WAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSN 160
A+ KG F G A R Y++ ++ +P N
Sbjct 139 LAQEEKNKGNEYFKKGDYPTAMRHYNEAVKRDPEN 173
> xla:447010 stip1, MGC82554; stress-induced-phosphoprotein 1
(Hsp70/Hsp90-organizing protein); K09553 stress-induced-phosphoprotein
1
Length=430
Score = 97.1 bits (240), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 61/191 (31%), Positives = 103/191 (53%), Gaps = 21/191 (10%)
Query 61 EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV 120
EEK KGN +FQ GD+ AV+H++ AI+ +P D LYSNR+ + L +F+ ALKD E C+
Sbjct 249 EEKNKGNESFQKGDYPQAVRHYSEAIKRNPNDAKLYSNRAACYTKLLEFQLALKDCEECI 308
Query 121 KLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLRDM 180
+L+P++ KGY+RK A + +A +Y K EL+ +++ + G + S + D
Sbjct 309 RLEPNFIKGYTRKAAALEAMKDYSKAMDAYQKATELDSTSKEAKDGYQRCMMSQYNRND- 367
Query 181 HALWAVSTALNRHPKLLK-YSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQPDV- 233
+P+ +K + DP + ++ +L Q+Q +P +L + P +
Sbjct 368 ------------NPEDVKRRAMADPEVQQIMSDPAMRLILEQMQKDPQALSEHLKNPVIA 415
Query 234 -RIREGMTVLL 243
RI++ M V L
Sbjct 416 QRIQKLMDVGL 426
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 69/142 (48%), Gaps = 11/142 (7%)
Query 56 QTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKD 115
+ QAQ+EK GN A++ DF A++H+ A +L P + +N++ + D+
Sbjct 109 KKQAQKEKELGNEAYKKKDFETALKHYGQAQELDPTNMTYITNQAAVYFEQADYNKCRDL 168
Query 116 AETCVKL----KPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
E +++ + D+ AK Y+R G + + + EA + ++K L + E L+K
Sbjct 169 CEKGIEVGRENREDYRQIAKAYARIGNSYYKEEKYKEATQFFNKSLAEHRTPEVLKK--- 225
Query 169 EVQQSGASLRDMHALWAVSTAL 190
QQ+ L++ L ++ L
Sbjct 226 -CQQAEKILKEQERLAYINPDL 246
> sce:YOR007C SGT2; Glutamine-rich cytoplasmic protein of unknown
function; contains tetratricopeptide (TPR) repeats, which
often mediate protein-protein interactions; has similarity
to human SGT, which is a cochaperone that negatively regulates
Hsp70
Length=346
Score = 89.0 bits (219), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 48/129 (37%), Positives = 82/129 (63%), Gaps = 8/129 (6%)
Query 53 DPAQTQAQEE--KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFE 110
D A+T+A+ E K +GN A N D+ A+ +T AI++ P + + Y+NR+ A +SLK+++
Sbjct 94 DDAETKAKAEDLKMQGNKAMANKDYELAINKYTEAIKVLPTNAIYYANRAAAHSSLKEYD 153
Query 111 NALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSN--EGLQKGL- 167
A+KDAE+ + + P + +GYSR G A++ G+ EA +Y K L++E N E +++
Sbjct 154 QAVKDAESAISIDPSYFRGYSRLGFAKYAQGKPEEALEAYKKVLDIEGDNATEAMKRDYE 213
Query 168 ---KEVQQS 173
K+V+QS
Sbjct 214 SAKKKVEQS 222
> xla:496358 sgta; small glutamine-rich tetratricopeptide repeat
(TPR)-containing, alpha
Length=302
Score = 82.8 bits (203), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 74/125 (59%), Gaps = 0/125 (0%)
Query 48 MATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLK 107
+A+ +D +A+ K +GN + +F +A+ ++T A++L+P + + Y NR+ A++ L
Sbjct 65 LASPSDEDLAEAERLKTEGNEQMKVENFESAISYYTKALELNPANAVYYCNRAAAYSKLG 124
Query 108 DFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGL 167
++ A++D E + + P+++K Y R GLA +L + EA Y + L L+P NE + L
Sbjct 125 NYAGAVRDCEAAITIDPNYSKAYGRMGLALSSLNKHAEAVGFYKQALVLDPDNETYKSNL 184
Query 168 KEVQQ 172
K +Q
Sbjct 185 KIAEQ 189
> dre:336867 fk20d10, wu:fa05b08, wu:fc52b05, wu:fk20d10, zgc:77080;
zgc:55741
Length=320
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 73/129 (56%), Gaps = 3/129 (2%)
Query 47 TMATAADPAQTQ---AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAF 103
T T P + Q A+ K GN + +F AAV+ ++ AIQL+PQ+ + + NR+ A+
Sbjct 76 TFVTTGSPYEHQLAEAERLKTDGNDQMKVENFSAAVEFYSKAIQLNPQNAVYFCNRAAAY 135
Query 104 ASLKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGL 163
+ L ++ A++D E + + +++K Y R GLA +L + EA Y K LEL+P N+
Sbjct 136 SKLGNYAGAVQDCERAIGIDANYSKAYGRMGLALASLNKYSEAVSYYKKALELDPDNDTY 195
Query 164 QKGLKEVQQ 172
+ L+ +Q
Sbjct 196 KVNLQVAEQ 204
> xla:414520 hypothetical protein MGC81394
Length=312
Score = 81.3 bits (199), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 74/125 (59%), Gaps = 0/125 (0%)
Query 48 MATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLK 107
+A+ +D +A+ K +GN + +F +AV ++T A++L+P++ + Y NR+ A++ L
Sbjct 77 LASPSDEDVAEAESLKTEGNEQMKVENFESAVTYYTKALELNPRNAVYYCNRAAAYSKLG 136
Query 108 DFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGL 167
++ A++D E + + P ++K Y R GLA +L + E+ Y + L L+P NE + L
Sbjct 137 NYAGAVRDCEEAISIDPSYSKAYGRMGLALSSLNKHAESVGFYKQALVLDPENETYKSNL 196
Query 168 KEVQQ 172
K +Q
Sbjct 197 KIAEQ 201
> ath:AT1G04190 tetratricopeptide repeat (TPR)-containing protein
Length=328
Score = 81.3 bits (199), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 68/128 (53%), Gaps = 3/128 (2%)
Query 48 MATAADPAQTQAQEEKA---KGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFA 104
MA A A + EK+ KGN F+ G+F A +T AI+L P + LYSNR+ AF
Sbjct 1 MAEKAGKATNGGEAEKSLKEKGNEFFKAGNFLKAAALYTQAIKLDPSNATLYSNRAAAFL 60
Query 105 SLKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQ 164
SL AL DAET +KL P W KGY RKG + + +A ++ L+ P + +
Sbjct 61 SLVKLSKALADAETTIKLNPQWEKGYFRKGCVLEAMEKYEDALAAFEMALQYNPQSTEVS 120
Query 165 KGLKEVQQ 172
+ +K + Q
Sbjct 121 RKIKRLGQ 128
> ath:AT3G04710 ankyrin repeat family protein
Length=455
Score = 80.9 bits (198), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 43/110 (39%), Positives = 60/110 (54%), Gaps = 0/110 (0%)
Query 65 KGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLKP 124
+G AF DF A+ +T AI P DH L+SNRS + L E+AL DA+ C +L P
Sbjct 333 RGQDAFHRKDFQMAIDAYTQAIDFDPTDHTLFSNRSLCWLRLGQAEHALSDAKACRELNP 392
Query 125 DWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSG 174
DW KG R+G A L R EA ++ +G+ L P ++ L +E +G
Sbjct 393 DWPKGCFREGAALRLLQRFDEAANAFYEGVLLSPESKELIDAFREAVDAG 442
> cpv:cgd2_1850 stress-induced protein sti1-like protein
Length=326
Score = 80.5 bits (197), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 92/180 (51%), Gaps = 14/180 (7%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A++ + +GN F+ ++ AA + + AI+ +P D LYSNR+ + L ++ +AL D +
Sbjct 140 AEKHRIEGNDLFKQKNYPAAKKEYDEAIKRNPSDSRLYSNRAACYMQLLEYPSALIDVQK 199
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
+ L P + K +SRKG + L +A +Y +GL+ +P N+ +GLK
Sbjct 200 ALDLDPKFTKAWSRKGNIHYFLKEYHKALHAYQEGLKCDPDNKECNEGLKNT-------- 251
Query 179 DMHALWAVSTALNRHPKLLKYSQQDPNYSKTLAK-----LLSQVQANPSSLRLVMAQPDV 233
M + VS++ + + ++ DP L+ +L Q++ NP++L V+ P +
Sbjct 252 -MAKIQQVSSSDQIDEEQVAHALADPEIQSLLSDPQFRLVLEQLKQNPATLTQVIQDPTI 310
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 57/120 (47%), Gaps = 14/120 (11%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVK- 121
K KGN ++ F A+ + AI++ P D +N+ + + +++ L E C++
Sbjct 9 KNKGNELYKQKKFDEALVQYDLAIEIDPNDISFLTNKGAVYLEMGEYQKCL---EVCMQA 65
Query 122 ------LKPDW---AKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
+K D+ AK Y+R + L +A+ Y K L LE +N + LKE+++
Sbjct 66 LEKRFEVKADFTKVAKAYNRMASCYIKMNELQKAKEMYEKSL-LEDNNRHTRTSLKELER 124
> mmu:218544 Sgtb, C630001O05Rik, MGC27660; small glutamine-rich
tetratricopeptide repeat (TPR)-containing, beta
Length=304
Score = 79.7 bits (195), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 39/115 (33%), Positives = 65/115 (56%), Gaps = 0/115 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+A + K +GN + ++ AAV +T AI+L P + + Y NR+ A + L + +A+KD E
Sbjct 84 KADQLKDEGNNHMKEENYAAAVDCYTQAIELDPNNAVYYCNRAAAQSKLSHYTDAIKDCE 143
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
+ + ++K Y R GLA + + EA SY K L+L+P N+ + LK +Q
Sbjct 144 KAIAIDSKYSKAYGRMGLALTAMNKFEEAVTSYQKALDLDPENDSYKSNLKIAEQ 198
> hsa:54557 SGTB, FLJ39002, SGT2; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, beta
Length=304
Score = 79.7 bits (195), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 65/115 (56%), Gaps = 0/115 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+A + K +GN + ++ AAV +T AI+L P + + Y NR+ A + L + +A+KD E
Sbjct 84 KADQLKDEGNNHMKEENYAAAVDCYTQAIELDPNNAVYYCNRAAAQSKLGHYTDAIKDCE 143
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
+ + ++K Y R GLA L + EA SY K L+L+P N+ + LK +Q
Sbjct 144 KAIAIDSKYSKAYGRMGLALTALNKFEEAVTSYQKALDLDPENDSYKSNLKIAEQ 198
> mmu:209683 Ttc28, 2310015L07Rik, 6030435N04, AI428795, AI851761,
BC002262, MGC7623; tetratricopeptide repeat domain 28
Length=2481
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 43/119 (36%), Positives = 68/119 (57%), Gaps = 2/119 (1%)
Query 53 DPAQTQAQ--EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFE 110
+PA ++A+ E+ + N A +GDFH A+ + A+ + PQ+ +LYSNRS A+ + +
Sbjct 44 EPALSKAEFVEKVRQSNQACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKTQQYH 103
Query 111 NALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE 169
AL DA L P W K Y R+G+A LGR +A +++ GL +P + L G+ E
Sbjct 104 KALDDAIKARLLNPKWPKAYFRQGVALQYLGRHADALAAFASGLAQDPKSLQLLVGMVE 162
> hsa:23331 TTC28, KIAA1043; tetratricopeptide repeat domain 28
Length=2481
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 40/109 (36%), Positives = 63/109 (57%), Gaps = 0/109 (0%)
Query 61 EEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCV 120
E+ + N A +GDFH A+ + A+ + PQ+ +LYSNRS A+ ++ ++ AL DA
Sbjct 60 EKVRQSNQACHDGDFHTAIVLYNEALAVDPQNCILYSNRSAAYMKIQQYDKALDDAIKAR 119
Query 121 KLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKE 169
L P W K Y R+G+A LGR +A +++ GL +P + L G+ E
Sbjct 120 LLNPKWPKAYFRQGVALQYLGRHADALAAFASGLAQDPKSLQLLVGMVE 168
> hsa:6449 SGTA, SGT, alphaSGT, hSGT; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=313
Score = 77.4 bits (189), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 68/123 (55%), Gaps = 8/123 (6%)
Query 54 PAQTQAQEE--------KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFAS 105
PA+T EE K +GN + +F AAV + AI+L+P + + + NR+ A++
Sbjct 78 PARTPPSEEDSAEAERLKTEGNEQMKVENFEAAVHFYGKAIELNPANAVYFCNRAAAYSK 137
Query 106 LKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQK 165
L ++ A++D E + + P ++K Y R GLA +L + EA Y K LEL+P NE +
Sbjct 138 LGNYAGAVQDCERAICIDPAYSKAYGRMGLALSSLNKHVEAVAYYKKALELDPDNETYKS 197
Query 166 GLK 168
LK
Sbjct 198 NLK 200
> hsa:79657 RPAP3, FLJ21908; RNA polymerase II associated protein
3
Length=631
Score = 77.0 bits (188), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 42/114 (36%), Positives = 63/114 (55%), Gaps = 0/114 (0%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K KGN F+ G + A+ +T + P + +L +NR+ A+ LK F A D V L
Sbjct 137 KEKGNKYFKQGKYDEAIDCYTKGMDADPYNPVLPTNRASAYFRLKKFAVAESDCNLAVAL 196
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGAS 176
+ K YSR+G A F L +L EA++ Y + LELEP+N L+++ Q+ AS
Sbjct 197 NRSYTKAYSRRGAARFALQKLEEAKKDYERVLELEPNNFEATNELRKISQALAS 250
Score = 65.9 bits (159), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 58/103 (56%), Gaps = 0/103 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
QA EK +GN F+ G + A++ +T I + LL +NR+ A+ ++ +E A KD
Sbjct 281 QAISEKDRGNGFFKEGKYERAIECYTRGIAADGANALLPANRAMAYLKIQKYEEAEKDCT 340
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSN 160
+ L ++K ++R+G A LG+L EA++ + L LEP N
Sbjct 341 QAILLDGSYSKAFARRGTARTFLGKLNEAKQDFETVLLLEPGN 383
> mmu:52551 Sgta, 5330427H01Rik, AI194281, D10Ertd190e, MGC6336,
Sgt, Stg; small glutamine-rich tetratricopeptide repeat (TPR)-containing,
alpha
Length=315
Score = 76.6 bits (187), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 39/123 (31%), Positives = 68/123 (55%), Gaps = 0/123 (0%)
Query 46 PTMATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFAS 105
P ++ +A+ K +GN + +F AAV + AI+L+P + + + NR+ A++
Sbjct 79 PDRTPPSEEDSAEAERLKTEGNEQMKLENFEAAVHLYGKAIELNPANAVYFCNRAAAYSK 138
Query 106 LKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQK 165
L ++ A++D E + + P ++K Y R GLA +L + EA Y K LEL+P N+ +
Sbjct 139 LGNYVGAVQDCERAIGIDPGYSKAYGRMGLALSSLNKHAEAVAYYKKALELDPDNDTYKS 198
Query 166 GLK 168
LK
Sbjct 199 NLK 201
> dre:431772 sgta, zgc:92462; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, alpha
Length=306
Score = 75.9 bits (185), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 46/163 (28%), Positives = 88/163 (53%), Gaps = 3/163 (1%)
Query 12 LLSNFFIFRGESSVLSVQTVFELLL--LCKLFFFSLPTMATAADPAQTQAQEEKAKGNAA 69
L + F I + + + Q + E+ L L K +LP + + + +A++ K +GN
Sbjct 39 LETTFKISSSDCHLAAPQPLREIFLNSLLKNDIVTLPKTFPSPEDIE-RAEQLKNEGNNH 97
Query 70 FQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLKPDWAKG 129
+ ++ +AV +T AI+L ++ + Y NR+ A + L+++ A+ D E + + P ++K
Sbjct 98 MKEENYSSAVDCYTKAIELDQRNAVYYCNRAAAHSKLENYTEAMGDCERAIAIDPSYSKA 157
Query 130 YSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
Y R GLA ++ + EA ++K L L+P N+ + LK V+Q
Sbjct 158 YGRMGLALTSMSKYPEAISYFNKALVLDPENDTYKSNLKIVEQ 200
> cel:R05F9.10 sgt-1; Small Glutamine-rich Tetratrico repeat protein
family member (sgt-1)
Length=337
Score = 74.7 bits (182), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 68/121 (56%), Gaps = 1/121 (0%)
Query 48 MATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLK 107
+ T +D +QA + K +GN + F AAVQ + AI+L+ +D + + NR+ A+ L+
Sbjct 94 LPTPSDSDISQANKLKEEGNDLMKASQFEAAVQKYNAAIKLN-RDPVYFCNRAAAYCRLE 152
Query 108 DFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGL 167
++ A++D T + L P ++K + R GLA R A +Y K LELEP+ E + L
Sbjct 153 QYDLAIQDCRTALALDPSYSKAWGRMGLAYSCQNRYEHAAEAYKKALELEPNQESYKNNL 212
Query 168 K 168
K
Sbjct 213 K 213
> dre:641500 MGC123010, wu:fk11h08; zgc:123010
Length=474
Score = 74.3 bits (181), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 37/93 (39%), Positives = 54/93 (58%), Gaps = 0/93 (0%)
Query 65 KGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLKP 124
KG Q G + AV FT AI+ P+D+ + NRS + L+ + AL DAE +++ P
Sbjct 190 KGIRFVQEGQYTQAVSLFTEAIKCDPKDYRFFGNRSYCYCCLEQYALALADAEKSIQMAP 249
Query 125 DWAKGYSRKGLAEFNLGRLGEAERSYSKGLELE 157
DW KGY R+G A L R EAE++ + L+L+
Sbjct 250 DWPKGYYRRGSALMGLKRYSEAEKAMEQVLKLD 282
> ath:AT4G22670 AtHip1; AtHip1 (Arabidopsis thaliana Hsp70-interacting
protein 1); binding
Length=441
Score = 73.9 bits (180), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 52/94 (55%), Gaps = 0/94 (0%)
Query 53 DPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENA 112
D + AQE K K A G+F A++H T AI L+P ++Y NR+ + LK A
Sbjct 117 DENREAAQEAKGKAMEALSEGNFDEAIEHLTRAITLNPTSAIMYGNRASVYIKLKKPNAA 176
Query 113 LKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEA 146
++DA +++ PD AKGY +G+A LG EA
Sbjct 177 IRDANAALEINPDSAKGYKSRGMARAMLGEWAEA 210
> hsa:79836 LONRF3, FLJ22612, MGC119463, MGC119465, RNF127; LON
peptidase N-terminal domain and ring finger 3
Length=759
Score = 73.2 bits (178), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 62/108 (57%), Gaps = 0/108 (0%)
Query 54 PAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENAL 113
P +A + + +GN ++ AA+ + A++L+P DHLLYSNRS + +L+ ENAL
Sbjct 238 PGPARASQLRHEGNRLYRERQVEAALLKYNEAVKLAPNDHLLYSNRSQIYFTLESHENAL 297
Query 114 KDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNE 161
DAE KL+P K + RK A LG++ EA R + + L+ N+
Sbjct 298 HDAEIACKLRPMGFKAHFRKAQALATLGKVEEALREFLYCVSLDGKNK 345
> xla:414472 rpap3, MGC81126; RNA polymerase II associated protein
3
Length=660
Score = 73.2 bits (178), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 46/142 (32%), Positives = 73/142 (51%), Gaps = 4/142 (2%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+A EK KGN F++G + A++ +T + P + +L +NR+ AF LK F A D
Sbjct 131 KALSEKEKGNNYFKSGKYDEAIECYTRGMDADPYNAILPTNRASAFFRLKKFAVAESDCN 190
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ----S 173
+ L D+AK Y+R+G A L L A+ Y K LEL+ +N + L+++ Q S
Sbjct 191 LAIALNRDYAKAYARRGAARLALKNLQGAKEDYEKVLELDANNFEAKNELRKINQELYSS 250
Query 174 GASLRDMHALWAVSTALNRHPK 195
+ +++ A A T N K
Sbjct 251 ASDVQENMATEAKITVENEEEK 272
Score = 57.0 bits (136), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 56/103 (54%), Gaps = 0/103 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
QA +K GNA F+ G + A++ ++ ++ + LL +NR+ A+ ++ ++ A D
Sbjct 283 QAIMQKDLGNAYFKEGKYEIAIECYSQGMEADNTNALLPANRAMAYLKIQKYKEAEADCT 342
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSN 160
+ L + K ++R+G A LG+ EA+ + L+L+P N
Sbjct 343 LAISLDASYCKAFARRGTASIMLGKQKEAKEDFEMVLKLDPGN 385
> xla:444591 sgtb, MGC84046; small glutamine-rich tetratricopeptide
repeat (TPR)-containing, beta
Length=308
Score = 72.8 bits (177), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 76/157 (48%), Gaps = 15/157 (9%)
Query 55 AQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALK 114
A +A++ K +GN + ++ AAV ++ AI+L P + + Y NR+ A + A+
Sbjct 85 AAEKAEQLKDEGNGLMKEQNYEAAVDCYSQAIELDPNNAVYYCNRAAAQSQRGKHSEAIT 144
Query 115 DAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ-- 172
D E + + ++K Y R G A + R EA SY K L+L+P NE + LK +Q
Sbjct 145 DCEKAISIDAKYSKAYGRMGRALVAMSRYKEAFESYQKALDLDPENESYRMNLKLAEQKL 204
Query 173 -------------SGASLRDMHALWAVSTALNRHPKL 196
ASL + A +++ +L R P++
Sbjct 205 RQIPSPISSEWGFDMASLMNNPAFVSMAASLMRDPQV 241
> mmu:71919 Rpap3, 2310042P20Rik, D15Ertd682e; RNA polymerase
II associated protein 3
Length=660
Score = 72.4 bits (176), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 38/111 (34%), Positives = 60/111 (54%), Gaps = 0/111 (0%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K KGN F+ G + A++ +T + P + +L +NR+ A+ LK F A D + L
Sbjct 138 KEKGNKYFKQGKYDEAIECYTKGMDADPYNPVLPTNRASAYFRLKKFAVAESDCNLAIAL 197
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQS 173
+ K Y+R+G A F L +L +A + Y K LELEP N L+++ Q+
Sbjct 198 SRTYTKAYARRGAARFALQKLEDARKDYEKVLELEPDNFEATNELRKINQA 248
Score = 63.2 bits (152), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 45/160 (28%), Positives = 77/160 (48%), Gaps = 11/160 (6%)
Query 50 TAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDF 109
T + +A EK GN F+ G + A++ +T I + LL +NR+ A+ ++ +
Sbjct 275 TGGQQGRQKAIAEKDLGNGFFKEGKYEQAIECYTRGIAADRTNALLPANRAMAYLKIQRY 334
Query 110 ENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNE----GLQK 165
E A +D + L ++K ++R+G A LG++ EA++ + L LEP N+ L +
Sbjct 335 EEAERDCTQAIVLDGSYSKAFARRGTARTFLGKINEAKQDFETVLLLEPGNKQAATELSR 394
Query 166 GLKEVQQSG-------ASLRDMHALWAVSTALNRHPKLLK 198
KE+ + G S + H + AV PK LK
Sbjct 395 IKKELIEKGHWDDVFLDSTQRHHVVKAVDNPPRGSPKALK 434
> mmu:74365 Lonrf3, 4932412G04Rik, 5730439E01Rik, A830039N02Rik,
AU023707, Rnf127; LON peptidase N-terminal domain and ring
finger 3
Length=753
Score = 71.2 bits (173), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/108 (36%), Positives = 62/108 (57%), Gaps = 0/108 (0%)
Query 54 PAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENAL 113
P +A + + +GN F+ AA+ + A++L+P DHLLYSNRS + +L+ E+AL
Sbjct 239 PGPARASQLRHEGNRLFREHQVEAALLKYNEAVRLAPNDHLLYSNRSQIYFTLESHEDAL 298
Query 114 KDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNE 161
DAE KL+P K + RK A LG++ EA + + + L+ N+
Sbjct 299 HDAEIACKLRPMGFKAHFRKAQALATLGKVKEALKEFLYCVSLDGKNK 346
> ath:AT3G17880 ATTDX; ATTDX (TETRATICOPEPTIDE DOMAIN-CONTAINING
THIOREDOXIN); oxidoreductase, acting on sulfur group of donors,
disulfide as acceptor / protein binding
Length=373
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 56/101 (55%), Gaps = 1/101 (0%)
Query 46 PTMATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFAS 105
PT A D + AQ EK+K A +G F A++H T A+ L+P +LY+ R+ F
Sbjct 93 PT-AEVTDENRDDAQSEKSKAMEAISDGRFDEAIEHLTKAVMLNPTSAILYATRASVFLK 151
Query 106 LKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEA 146
+K A++DA ++ D AKGY +G+A+ LG+ EA
Sbjct 152 VKKPNAAIRDANVALQFNSDSAKGYKSRGMAKAMLGQWEEA 192
> ath:AT4G08320 tetratricopeptide repeat (TPR)-containing protein
Length=427
Score = 70.9 bits (172), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 37/115 (32%), Positives = 69/115 (60%), Gaps = 1/115 (0%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A+ K +GN A Q+ + AV+ ++ AI L+ ++ + Y NR+ A+ + A+KD
Sbjct 175 AETLKCQGNKAMQSNLYLEAVELYSFAIALTDKNAVFYCNRAAAYTQINMCSEAIKDCLK 234
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEA-ERSYSKGLELEPSNEGLQKGLKEVQQ 172
+++ P+++K YSR GLA + G+ EA E+ + K L L+P NE +++ ++ +Q
Sbjct 235 SIEIDPNYSKAYSRLGLAYYAQGKYAEAIEKGFKKALLLDPHNESVKENIRVAEQ 289
> cel:T12D8.8 hypothetical protein; K09560 suppressor of tumorigenicity
protein 13
Length=422
Score = 70.1 bits (170), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 40/124 (32%), Positives = 68/124 (54%), Gaps = 1/124 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
+A EE+ K AF NGDF A+ HFT AI+ +P +L++ R+ LK A+ D +
Sbjct 114 KASEERGKAQEAFSNGDFDTALTHFTAAIEANPGSAMLHAKRANVLLKLKRPVAAIADCD 173
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASL 177
+ + PD A+GY +G A LG+ EA+ + +L+ +E + LKEV+ + +
Sbjct 174 KAISINPDSAQGYKFRGRANRLLGKWVEAKTDLATACKLD-YDEAANEWLKEVEPNAHKI 232
Query 178 RDMH 181
++ +
Sbjct 233 QEYN 236
> dre:560112 si:ch211-243o19.6, wu:fc10f01; si:dkey-33c12.4
Length=631
Score = 70.1 bits (170), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 38/115 (33%), Positives = 65/115 (56%), Gaps = 0/115 (0%)
Query 57 TQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDA 116
T++ E GN +G+ AV++FT+AI+ +P+++ L+ NRS + + +E +L DA
Sbjct 296 TRSVELAVIGNEYAGSGNMEMAVKYFTDAIKHNPKEYKLFGNRSYCYEKMLQYEKSLTDA 355
Query 117 ETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQ 171
E + + P W KG RKG A L R EA ++ + L+L+ S + + + VQ
Sbjct 356 EIALSMNPKWIKGLYRKGRALVGLKRYNEARLTFGEVLKLDSSCKDAAEEIMRVQ 410
> xla:398004 ppp5c, pp5; protein phosphatase 5, catalytic subunit
(EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=493
Score = 69.7 bits (169), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 61/114 (53%), Gaps = 0/114 (0%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A+E K + N F+ D+ AVQ++T AI LSP + Y NRS A+ + + AL DA
Sbjct 22 AEELKEQANEYFRVKDYDHAVQYYTQAIDLSPDTAIYYGNRSLAYLRTECYGYALADASR 81
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQ 172
++L + KGY R+ + LG+L A + Y +++ P ++ Q +E +
Sbjct 82 AIQLDAKYIKGYYRRAASNMALGKLKAALKDYETVVKVRPHDKDAQMKFQECNK 135
> ath:AT2G42810 PP5.2; PP5.2 (PROTEIN PHOSPHATASE 5.2); phosphoprotein
phosphatase/ protein binding / protein serine/threonine
phosphatase; K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=538
Score = 68.9 bits (167), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 74/122 (60%), Gaps = 0/122 (0%)
Query 57 TQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDA 116
++A+E K++ N AF+ + +A+ +T AI+L+ + + ++NR+ A L+++ +A++DA
Sbjct 11 SRAEEFKSQANEAFKGHKYSSAIDLYTKAIELNSNNAVYWANRAFAHTKLEEYGSAIQDA 70
Query 117 ETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGAS 176
+++ ++KGY R+G A +G+ +A + + + L P++ + LKE +++
Sbjct 71 SKAIEVDSRYSKGYYRRGAAYLAMGKFKDALKDFQQVKRLSPNDPDATRKLKECEKAVMK 130
Query 177 LR 178
L+
Sbjct 131 LK 132
> mmu:56354 Dnajc7, 2010003F24Rik, 2010004G07Rik, CCRP, Ttc2,
mDj11, mTpr2; DnaJ (Hsp40) homolog, subfamily C, member 7; K09527
DnaJ homolog subfamily C member 7
Length=494
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 37/116 (31%), Positives = 61/116 (52%), Gaps = 0/116 (0%)
Query 53 DPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENA 112
+ A+ +A+ K +GNA + D++ A ++T AI + P + Y NR+ L F A
Sbjct 22 EDAKREAESFKEQGNAYYAKKDYNEAYNYYTKAIDMCPNNASYYGNRAATLMMLGRFREA 81
Query 113 LKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLK 168
L DA+ V+L + +G+ R+G +LG A RS+ + LEL+ N Q+ K
Sbjct 82 LGDAQQSVRLDDSFVRGHLREGKCHLSLGNAMAACRSFQRALELDHKNAQAQQEFK 137
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 52/107 (48%), Gaps = 4/107 (3%)
Query 62 EKAKGNAAFQNGDFHAAVQHFTNAIQLSPQD----HLLYSNRSGAFASLKDFENALKDAE 117
+K GN AF+ G++ A + +T A+ + P + LY NR + L+ E+A++D
Sbjct 259 KKEDGNKAFKEGNYKLAYELYTEALGIDPNNIKTNAKLYCNRGTVNSKLRQLEDAIEDCT 318
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQ 164
VKL + K Y R+ + + EA R Y K + E + E Q
Sbjct 319 NAVKLDDTYIKAYLRRAQCYMDTEQFEEAVRDYEKVYQTEKTKEHKQ 365
> hsa:6674 SPAG1, FLJ32920, HSD-3.8, SP75, TPIS; sperm associated
antigen 1
Length=926
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 65/115 (56%), Gaps = 1/115 (0%)
Query 59 AQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAET 118
A EK KGN AF +GD+ AV ++T +I P + Y+NR+ A L+++ +A +D E
Sbjct 209 ATREKEKGNEAFNSGDYEEAVMYYTRSISALPT-VVAYNNRAQAEIKLQNWNSAFQDCEK 267
Query 119 CVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQS 173
++L+P K R+ + +L EA SK L++EP N+ +K L EV++
Sbjct 268 VLELEPGNVKALLRRATTYKHQNKLREATEDLSKVLDVEPDNDLAKKTLSEVERD 322
Score = 38.9 bits (89), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 22/97 (22%), Positives = 50/97 (51%), Gaps = 0/97 (0%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K +GN + ++ A+ ++ ++++ ++ +Y+NR+ + L FE A +D + ++L
Sbjct 627 KEEGNQCVNDKNYKDALSKYSECLKINNKECAIYTNRALCYLKLCQFEEAKQDCDQALQL 686
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPS 159
K + R+ LA L ++ +K + L+PS
Sbjct 687 ADGNVKAFYRRALAHKGLKNYQKSLIDLNKVILLDPS 723
Score = 36.2 bits (82), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 49/103 (47%), Gaps = 8/103 (7%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAI--------QLSPQDHLLYSNRSGAFASLKDFENALK 114
K++GN F++G F A ++ AI +++ +LYSNR+ + + ++
Sbjct 449 KSQGNELFRSGQFAEAAGKYSAAIALLEPAGSEIADDLSILYSNRAACYLKEGNCSGCIQ 508
Query 115 DAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELE 157
D ++L P K R+ +A L + G+A Y L+++
Sbjct 509 DCNRALELHPFSMKPLLRRAMAYETLEQYGKAYVDYKTVLQID 551
> hsa:5536 PPP5C, FLJ36922, FLJ55954, PP5, PPP5, PPT; protein
phosphatase 5, catalytic subunit (EC:3.1.3.16); K04460 protein
phosphatase 5 [EC:3.1.3.16]
Length=477
Score = 66.6 bits (161), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 36/127 (28%), Positives = 67/127 (52%), Gaps = 0/127 (0%)
Query 46 PTMATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFAS 105
P AD A +A+E K + N F+ D+ A++ ++ AI+L+P + + Y NRS A+
Sbjct 15 PRDEPPADGALKRAEELKTQANDYFKAKDYENAIKFYSQAIELNPSNAIYYGNRSLAYLR 74
Query 106 LKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQK 165
+ + AL DA ++L + KGY R+ + LG+ A R Y ++++P ++ +
Sbjct 75 TECYGYALGDATRAIELDKKYIKGYYRRAASNMALGKFRAALRDYETVVKVKPHDKDAKM 134
Query 166 GLKEVQQ 172
+E +
Sbjct 135 KYQECNK 141
> hsa:64427 TTC31, FLJ12788, FLJ33201, MGC120200; tetratricopeptide
repeat domain 31
Length=519
Score = 66.2 bits (160), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 38/89 (42%), Positives = 49/89 (55%), Gaps = 0/89 (0%)
Query 58 QAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAE 117
Q+QE G + QNG +H AV FT A++L+PQDH L+ NRS L AL DA+
Sbjct 304 QSQELAKLGTSFAQNGFYHEAVVLFTQALKLNPQDHRLFGNRSFCHERLGQPAWALADAQ 363
Query 118 TCVKLKPDWAKGYSRKGLAEFNLGRLGEA 146
+ L+P W +G R G A L R EA
Sbjct 364 VALTLRPGWPRGLFRLGKALMGLQRFREA 392
> mmu:19060 Ppp5c, AU020526, PP5; protein phosphatase 5, catalytic
subunit (EC:3.1.3.16); K04460 protein phosphatase 5 [EC:3.1.3.16]
Length=499
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/116 (29%), Positives = 63/116 (54%), Gaps = 0/116 (0%)
Query 46 PTMATAADPAQTQAQEEKAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFAS 105
P AD +A+E K + N F+ D+ A++ ++ AI+L+P + + Y NRS A+
Sbjct 15 PRDEPPADGTLKRAEELKTQANDYFKAKDYENAIKFYSQAIELNPGNAIYYGNRSLAYLR 74
Query 106 LKDFENALKDAETCVKLKPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNE 161
+ + AL DA ++L + KGY R+ + LG+ A R Y ++++P+++
Sbjct 75 TECYGYALGDATRAIELDKKYIKGYYRRAASNMALGKFRAALRDYETVVKVKPNDK 130
> mmu:22129 Ttc3, 2610202A04Rik, AA409221, D16Ium21, D16Ium21e,
KIAA4119, TPRD, mKIAA4119; tetratricopeptide repeat domain
3
Length=1979
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 60/116 (51%), Gaps = 3/116 (2%)
Query 63 KAKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKL 122
K +GN F F AV ++T AI+ P++HLLY NR+ F + F NAL D + + L
Sbjct 235 KMRGNEEFSKEKFEIAVIYYTRAIEYRPENHLLYGNRALCFLRMGQFRNALSDGKRAIVL 294
Query 123 KPDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQSGASLR 178
K W KG+ R A L LGE + + ++ + + +G+K++ Q L+
Sbjct 295 KNTWPKGHYRYCDA---LCMLGEYDWALQANIKAQKLCKNDPEGIKDLIQQHVKLQ 347
> dre:393286 ttc25, MGC56362, zgc:56362; tetratricopeptide repeat
domain 25
Length=486
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 37/110 (33%), Positives = 62/110 (56%), Gaps = 0/110 (0%)
Query 64 AKGNAAFQNGDFHAAVQHFTNAIQLSPQDHLLYSNRSGAFASLKDFENALKDAETCVKLK 123
A+G+ FQ G++ AV+ FT A+ L P + +RS + L D ENALKDAE+ +K
Sbjct 19 AEGDQLFQRGEYVKAVESFTTALTLQPDNKNCLVSRSRCYVKLGDAENALKDAESSLKDN 78
Query 124 PDWAKGYSRKGLAEFNLGRLGEAERSYSKGLELEPSNEGLQKGLKEVQQS 173
++ KG +K A + +G A Y +G +L P + + G+++ Q++
Sbjct 79 KNYFKGLYQKAEALYTMGDFEFALVYYHRGHKLRPELQEFRLGIQKAQEA 128
Lambda K H
0.318 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8594937416
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40