bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2724_orf1
Length=123
Score E
Sequences producing significant alignments: (Bits) Value
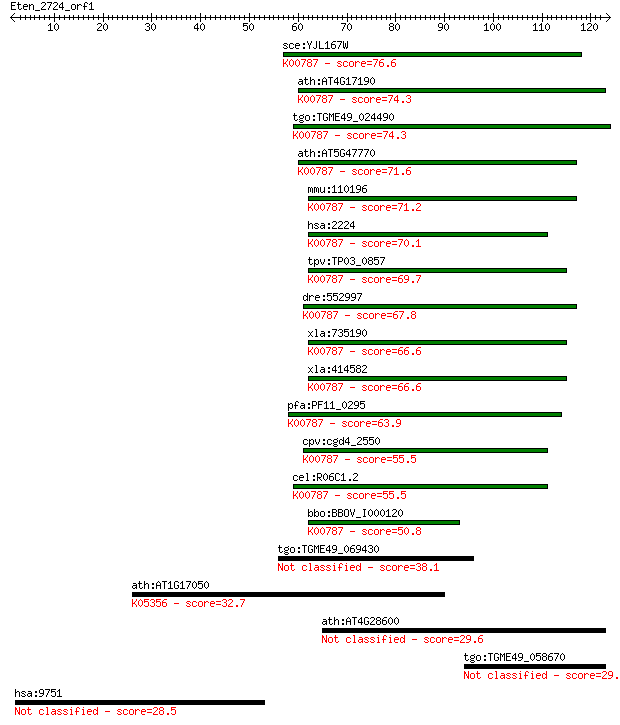
sce:YJL167W ERG20, BOT3, FDS1, FPP1; Erg20p (EC:2.5.1.10 2.5.1... 76.6 2e-14
ath:AT4G17190 FPS2; FPS2 (FARNESYL DIPHOSPHATE SYNTHASE 2); di... 74.3 8e-14
tgo:TGME49_024490 farnesyl pyrophosphate synthetase, putative ... 74.3 9e-14
ath:AT5G47770 FPS1; FPS1 (FARNESYL DIPHOSPHATE SYNTHASE 1); di... 71.6 5e-13
mmu:110196 Fdps, 6030492I17Rik, AI256750, Fdpsl1, MGC107162, m... 71.2 7e-13
hsa:2224 FDPS, FPPS, FPS; farnesyl diphosphate synthase (EC:2.... 70.1 2e-12
tpv:TP03_0857 farnesyl pyrophosphate synthetase (EC:2.5.1.1); ... 69.7 2e-12
dre:552997 fdps, MGC114048, im:6904202, wu:fb23g06; farnesyl d... 67.8 8e-12
xla:735190 fdps, MGC131256; farnesyl diphosphate synthase; K00... 66.6 2e-11
xla:414582 hypothetical protein MGC83119; K00787 farnesyl diph... 66.6 2e-11
pfa:PF11_0295 farnesyl pyrophosphate synthase, putative (EC:2.... 63.9 1e-10
cpv:cgd4_2550 farnesyl pyrophosphate synthase ; K00787 farnesy... 55.5 4e-08
cel:R06C1.2 hypothetical protein; K00787 farnesyl diphosphate ... 55.5 5e-08
bbo:BBOV_I000120 16.m00760; farnesyl pyrophosphate synthetase ... 50.8 1e-06
tgo:TGME49_069430 hexaprenyl pyrophosphate synthetase protein,... 38.1 0.006
ath:AT1G17050 SPS2; SPS2 (Solanesyl diphosphate synthase 2); t... 32.7 0.29
ath:AT4G28600 NPGR2; NPGR2 (no pollen germination related 2); ... 29.6 2.7
tgo:TGME49_058670 hypothetical protein 29.3 3.0
hsa:9751 SNPH, KIAA0374, MGC46096, bA314N13.5; syntaphilin 28.5 6.1
> sce:YJL167W ERG20, BOT3, FDS1, FPP1; Erg20p (EC:2.5.1.10 2.5.1.1);
K00787 farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=352
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 36/64 (56%), Positives = 45/64 (70%), Gaps = 4/64 (6%)
Query 57 SELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL---IYS 113
++ +LGW +ELLQA FLVADD MD + TRRG+ CWY+ PEVG AINDA L IY
Sbjct 80 EKVAILGWCIELLQAYFLVADDMMDKSITRRGQPCWYKVPEVGEI-AINDAFMLEAAIYK 138
Query 114 VHQS 117
+ +S
Sbjct 139 LLKS 142
> ath:AT4G17190 FPS2; FPS2 (FARNESYL DIPHOSPHATE SYNTHASE 2);
dimethylallyltranstransferase/ geranyltranstransferase; K00787
farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=342
Score = 74.3 bits (181), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 32/63 (50%), Positives = 41/63 (65%), Gaps = 1/63 (1%)
Query 60 CVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQSDP 119
C LGW +E LQA FLV DD MD + TRRG+ CW+R+P+VG AIND + L +H+
Sbjct 76 CALGWCIEWLQAYFLVLDDIMDNSVTRRGQPCWFRKPKVGMI-AINDGILLRNHIHRILK 134
Query 120 PSF 122
F
Sbjct 135 KHF 137
> tgo:TGME49_024490 farnesyl pyrophosphate synthetase, putative
(EC:2.5.1.1); K00787 farnesyl diphosphate synthase [EC:2.5.1.1
2.5.1.10]
Length=646
Score = 74.3 bits (181), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 36/71 (50%), Positives = 47/71 (66%), Gaps = 6/71 (8%)
Query 59 LCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL---IYSV- 114
L LGW VELLQ+ FLV DD MD + TRRGK CWYR +G +NA+ND++ L +Y V
Sbjct 313 LAALGWCVELLQSCFLVMDDVMDHSLTRRGKQCWYRCDGIGVSNAVNDSLVLEAAVYRVL 372
Query 115 --HQSDPPSFA 123
+ D P++A
Sbjct 373 REYLGDHPAYA 383
> ath:AT5G47770 FPS1; FPS1 (FARNESYL DIPHOSPHATE SYNTHASE 1);
dimethylallyltranstransferase/ geranyltranstransferase; K00787
farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=384
Score = 71.6 bits (174), Expect = 5e-13, Method: Composition-based stats.
Identities = 31/57 (54%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Query 60 CVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQ 116
C LGW +E LQA FLV DD MD + TRRG+ CW+R P+VG AIND + L +H+
Sbjct 118 CALGWCIEWLQAYFLVLDDIMDNSVTRRGQPCWFRVPQVGMV-AINDGILLRNHIHR 173
> mmu:110196 Fdps, 6030492I17Rik, AI256750, Fdpsl1, MGC107162,
mKIAA1293; farnesyl diphosphate synthetase (EC:2.5.1.1 2.5.1.10);
K00787 farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=353
Score = 71.2 bits (173), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 31/55 (56%), Positives = 43/55 (78%), Gaps = 1/55 (1%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQ 116
+GW VELLQA FLV+DD MD + TRRG+ CWY++P +G +AINDA+ L S+++
Sbjct 88 VGWCVELLQAFFLVSDDIMDSSLTRRGQICWYQKPGIG-LDAINDALLLEASIYR 141
> hsa:2224 FDPS, FPPS, FPS; farnesyl diphosphate synthase (EC:2.5.1.1
2.5.1.10); K00787 farnesyl diphosphate synthase [EC:2.5.1.1
2.5.1.10]
Length=419
Score = 70.1 bits (170), Expect = 2e-12, Method: Composition-based stats.
Identities = 32/49 (65%), Positives = 38/49 (77%), Gaps = 1/49 (2%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+GW VELLQA FLVADD MD + TRRG+ CWY++P VG +AINDA L
Sbjct 154 VGWCVELLQAFFLVADDIMDSSLTRRGQICWYQKPGVG-LDAINDANLL 201
> tpv:TP03_0857 farnesyl pyrophosphate synthetase (EC:2.5.1.1);
K00787 farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=353
Score = 69.7 bits (169), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 29/56 (51%), Positives = 36/56 (64%), Gaps = 3/56 (5%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVF---LIYSV 114
+ W VELLQ +FLVADD +D + RR CWY P +G NAIND +F LIY +
Sbjct 90 MSWCVELLQTSFLVADDIIDKSLKRRSNTCWYLVPTIGVENAINDVMFLYTLIYRI 145
> dre:552997 fdps, MGC114048, im:6904202, wu:fb23g06; farnesyl
diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase,
geranyltranstransferase); K00787
farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=356
Score = 67.8 bits (164), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 30/56 (53%), Positives = 42/56 (75%), Gaps = 1/56 (1%)
Query 61 VLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYSVHQ 116
++GW +ELLQA FLVADD MD + TRRG+ CWY++ +G +AINDA L S+++
Sbjct 90 LVGWCIELLQAFFLVADDIMDSSVTRRGQPCWYKKEAIG-LDAINDAFLLEGSIYR 144
> xla:735190 fdps, MGC131256; farnesyl diphosphate synthase; K00787
farnesyl diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=348
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/56 (55%), Positives = 40/56 (71%), Gaps = 4/56 (7%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL---IYSV 114
+GW VELLQA FLVADD MD + TRRG+ CWYR+ +G +A+ND+ L IY +
Sbjct 83 VGWCVELLQAFFLVADDIMDNSVTRRGQPCWYRKEGIG-LDAVNDSFLLEAGIYRI 137
> xla:414582 hypothetical protein MGC83119; K00787 farnesyl diphosphate
synthase [EC:2.5.1.1 2.5.1.10]
Length=348
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/56 (55%), Positives = 40/56 (71%), Gaps = 4/56 (7%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL---IYSV 114
+GW VELLQA FLVADD MD + TRRG+ CWYR+ +G +A+ND+ L IY +
Sbjct 83 VGWCVELLQAFFLVADDIMDNSVTRRGQPCWYRKEGIG-LDAVNDSFLLEACIYRI 137
> pfa:PF11_0295 farnesyl pyrophosphate synthase, putative (EC:2.5.1.10);
K00787 farnesyl diphosphate synthase [EC:2.5.1.1
2.5.1.10]
Length=376
Score = 63.9 bits (154), Expect = 1e-10, Method: Composition-based stats.
Identities = 29/56 (51%), Positives = 39/56 (69%), Gaps = 1/56 (1%)
Query 58 ELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFLIYS 113
++ + W +E+LQA+FLVADD MD TRR K CWY +V NA+ND VFL+Y+
Sbjct 87 KVACIAWCIEILQASFLVADDIMDKGETRRNKHCWYLLKDVEIKNAVND-VFLLYN 141
> cpv:cgd4_2550 farnesyl pyrophosphate synthase ; K00787 farnesyl
diphosphate synthase [EC:2.5.1.1 2.5.1.10]
Length=384
Score = 55.5 bits (132), Expect = 4e-08, Method: Composition-based stats.
Identities = 27/50 (54%), Positives = 31/50 (62%), Gaps = 2/50 (4%)
Query 61 VLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+LGW VE +QA L+ADD MD RRG CWY G +NAIND FL
Sbjct 115 LLGWVVEAIQALILIADDIMDSGKFRRGAPCWYIVH--GQSNAINDIFFL 162
> cel:R06C1.2 hypothetical protein; K00787 farnesyl diphosphate
synthase [EC:2.5.1.1 2.5.1.10]
Length=352
Score = 55.5 bits (132), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 25/52 (48%), Positives = 36/52 (69%), Gaps = 1/52 (1%)
Query 59 LCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRRPEVGPANAINDAVFL 110
+C +E++Q+ +L+ADD MD + TRRGK CW+RR VG +AINDA +
Sbjct 81 VCEAAATLEIIQSFYLIADDIMDNSETRRGKPCWFRREGVG-MSAINDAFIM 131
> bbo:BBOV_I000120 16.m00760; farnesyl pyrophosphate synthetase
(EC:2.5.1.1); K00787 farnesyl diphosphate synthase [EC:2.5.1.1
2.5.1.10]
Length=336
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 20/31 (64%), Positives = 22/31 (70%), Gaps = 0/31 (0%)
Query 62 LGWAVELLQAAFLVADDQMDGAFTRRGKACW 92
+GW VELLQ AFLVADD MD + RR CW
Sbjct 92 MGWCVELLQTAFLVADDIMDKSVMRRSNLCW 122
> tgo:TGME49_069430 hexaprenyl pyrophosphate synthetase protein,
putative (EC:2.5.1.30)
Length=536
Score = 38.1 bits (87), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/40 (50%), Positives = 27/40 (67%), Gaps = 1/40 (2%)
Query 56 GSELCVLGWAVELLQAAFLVADDQMDGAFTRRGKACWYRR 95
G ELC++ A EL+ A L+ DD +DGA TRRG+ +RR
Sbjct 233 GLELCMVHVA-ELIHTASLMHDDVIDGADTRRGQPATHRR 271
> ath:AT1G17050 SPS2; SPS2 (Solanesyl diphosphate synthase 2);
trans-octaprenyltranstransferase; K05356 all-trans-nonaprenyl-diphosphate
synthase [EC:2.5.1.84 2.5.1.85]
Length=417
Score = 32.7 bits (73), Expect = 0.29, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 17/81 (20%)
Query 26 VTISGGRNPAAPGSADDSFAAG-------------EAALDKAFGSELCV----LGWAVEL 68
++I G NP +A+ F+AG A + A EL V LG +E+
Sbjct 114 LSIVGAENPVLISAAEQIFSAGGKRMRPGLVFLVSRATAELAGLKELTVEHRRLGEIIEM 173
Query 69 LQAAFLVADDQMDGAFTRRGK 89
+ A L+ DD +D + RRG+
Sbjct 174 IHTASLIHDDVLDESDMRRGR 194
> ath:AT4G28600 NPGR2; NPGR2 (no pollen germination related 2);
calmodulin binding
Length=739
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 20/60 (33%), Positives = 27/60 (45%), Gaps = 2/60 (3%)
Query 65 AVELLQAAFLVADDQMDGAFTRRGKAC--WYRRPEVGPANAINDAVFLIYSVHQSDPPSF 122
AVELL + +AD D + R W PE AVFL+YS ++ PP+
Sbjct 217 AVELLPELWKLADSPRDAILSYRRALLNHWKLDPETTARIQKEYAVFLLYSGEEAVPPNL 276
> tgo:TGME49_058670 hypothetical protein
Length=1714
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 15/31 (48%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 94 RRPE-VGPANAINDAVFLIYSVHQSD-PPSF 122
RRPE GP D++FL+ + +SD PPSF
Sbjct 1579 RRPEEPGPLEKAKDSLFLLGEIEESDVPPSF 1609
> hsa:9751 SNPH, KIAA0374, MGC46096, bA314N13.5; syntaphilin
Length=494
Score = 28.5 bits (62), Expect = 6.1, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 26/53 (49%), Gaps = 2/53 (3%)
Query 2 TSAAAAAAPAFSQTGATCGTVARPVTISGGRNPAAP--GSADDSFAAGEAALD 52
T +A +PA S T ++ T + G R P P GSA+D +G AA D
Sbjct 197 TGESAGGSPARSLTRSSTYTKLSDPAVCGDRQPGDPSSGSAEDGADSGFAAAD 249
Lambda K H
0.318 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2003197800
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40