bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2713_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
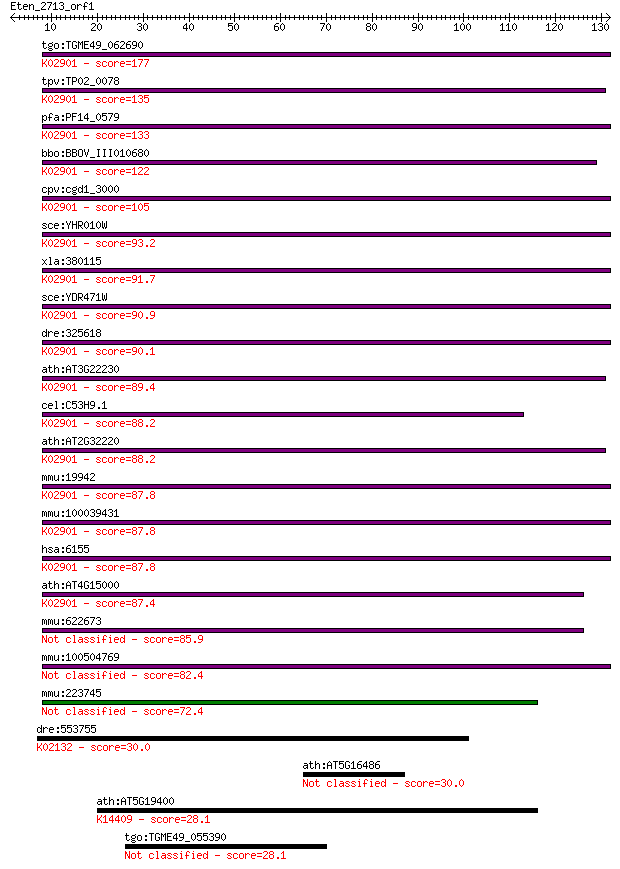
tgo:TGME49_062690 ribosomal protein L27, putative ; K02901 lar... 177 1e-44
tpv:TP02_0078 60S ribosomal protein L27e; K02901 large subunit... 135 4e-32
pfa:PF14_0579 60S ribosomal protein L27, putative; K02901 larg... 133 1e-31
bbo:BBOV_III010680 17.m07920; ribosomal L27e protein family pr... 122 2e-28
cpv:cgd1_3000 60S ribosomal protein L27 ; K02901 large subunit... 105 4e-23
sce:YHR010W RPL27A, RPL27; L27A; K02901 large subunit ribosoma... 93.2 2e-19
xla:380115 rpl27, MGC68687; ribosomal protein L27 (EC:3.6.5.3)... 91.7 6e-19
sce:YDR471W RPL27B; L27B; K02901 large subunit ribosomal prote... 90.9 1e-18
dre:325618 rpl27, fc93a08, wu:fc93a08, zgc:56171; ribosomal pr... 90.1 2e-18
ath:AT3G22230 60S ribosomal protein L27 (RPL27B); K02901 large... 89.4 3e-18
cel:C53H9.1 rpl-27; Ribosomal Protein, Large subunit family me... 88.2 5e-18
ath:AT2G32220 60S ribosomal protein L27 (RPL27A); K02901 large... 88.2 6e-18
mmu:19942 Rpl27, MGC103134; ribosomal protein L27 (EC:3.6.5.3)... 87.8 7e-18
mmu:100039431 Gm11518; 60S ribosomal protein L27; K02901 large... 87.8 7e-18
hsa:6155 RPL27; ribosomal protein L27; K02901 large subunit ri... 87.8 7e-18
ath:AT4G15000 60S ribosomal protein L27 (RPL27C); K02901 large... 87.4 9e-18
mmu:622673 Gm6341, EG622673; predicted pseudogene 6341 85.9
mmu:100504769 60S ribosomal protein L27-like 82.4 3e-16
mmu:223745 Gm4825, EG223745; predicted pseudogene 4825 72.4
dre:553755 atp5a1, zgc:154103; ATP synthase, H+ transporting, ... 30.0 1.9
ath:AT5G16486 hypothetical protein 30.0 2.0
ath:AT5G19400 SMG7; SMG7; K14409 protein SMG7 28.1 7.2
tgo:TGME49_055390 hypothetical protein 28.1 8.0
> tgo:TGME49_062690 ribosomal protein L27, putative ; K02901 large
subunit ribosomal protein L27e
Length=146
Score = 177 bits (448), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 84/124 (67%), Positives = 99/124 (79%), Gaps = 0/124 (0%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKR 67
LK GRVVVVLNGR AGKKGVVV+T EG+KER F +CLVAGIEKAPL+V K+M K++KR
Sbjct 5 LKSGRVVVVLNGRHAGKKGVVVNTWEGSKERQFAFCLVAGIEKAPLKVHKKMPAKKIEKR 64
Query 68 MRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFMN 127
MR K F++Y+NV HLMPTRYTVS+D DVK + PE ++ RK+AKK L IFYEKF N
Sbjct 65 MRVKPFLKYINVNHLMPTRYTVSSDMDVKTMVPEDAMRTVDDRKAAKKGLKKIFYEKFTN 124
Query 128 PVNE 131
PVNE
Sbjct 125 PVNE 128
> tpv:TP02_0078 60S ribosomal protein L27e; K02901 large subunit
ribosomal protein L27e
Length=148
Score = 135 bits (339), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 64/124 (51%), Positives = 93/124 (75%), Gaps = 1/124 (0%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVSTSEGT-KERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LKPGRVV++L+GR AG K VVV T+E + K+R + CLVAG+EKAP++V+K+MS+ K++K
Sbjct 5 LKPGRVVILLSGRRAGCKAVVVQTNESSSKKRPYLNCLVAGVEKAPMKVTKKMSSKKIEK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R++ KAF++YVNV HLMPTRY V+ D K+L + +E+ + RK A+K++ + E F
Sbjct 65 RLKLKAFVKYVNVNHLMPTRYMVTTTLDPKSLVSDEQMENKSSRKEARKSVKAVLEECFS 124
Query 127 NPVN 130
NP N
Sbjct 125 NPEN 128
> pfa:PF14_0579 60S ribosomal protein L27, putative; K02901 large
subunit ribosomal protein L27e
Length=146
Score = 133 bits (335), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 69/125 (55%), Positives = 95/125 (76%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVSTSEG-TKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LKPG+V+++LNGR AGKK V+V+T EG T+ER + YCLVAGIEK PL+V+K M+ K+ K
Sbjct 5 LKPGKVIIILNGRRAGKKAVIVNTYEGQTRERPYSYCLVAGIEKHPLKVNKSMTKKKIVK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + KAFI+ +NV H++PTRY V+NDFD+K+LA + L+ S +K K L IF +KF+
Sbjct 65 RSKVKAFIKCINVNHILPTRYQVANDFDIKSLASDDVLK-SKNKKKEVKKLGKIFRDKFL 123
Query 127 NPVNE 131
PVN+
Sbjct 124 EPVNK 128
> bbo:BBOV_III010680 17.m07920; ribosomal L27e protein family
protein; K02901 large subunit ribosomal protein L27e
Length=146
Score = 122 bits (307), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 57/122 (46%), Positives = 84/122 (68%), Gaps = 1/122 (0%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVSTSEG-TKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPGRVV+VL GR AG+KG++V +E +K R + CL+AGI++ P +V+++MS +++K
Sbjct 5 MKPGRVVIVLTGRRAGRKGIIVQCNENVSKRRPYANCLIAGIDRYPSKVTRKMSKEQIKK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R+R K F++YVNV HLMPTRY V+ D KAL ++ +E RK A+ AL +F E
Sbjct 65 RLRIKPFVKYVNVNHLMPTRYVVTGRLDPKALVTDSQMESRETRKEARMALKAVFEEALN 124
Query 127 NP 128
P
Sbjct 125 TP 126
> cpv:cgd1_3000 60S ribosomal protein L27 ; K02901 large subunit
ribosomal protein L27e
Length=162
Score = 105 bits (261), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 63/125 (50%), Positives = 89/125 (71%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVSTSE-GTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+K GRVVV+LNGR AGKK VVV+T E GTK+R F + LVAG+EKAPL+V KR+S K++K
Sbjct 22 MKQGRVVVLLNGRYAGKKAVVVNTFESGTKDRPFPFVLVAGVEKAPLKVHKRLSKEKLKK 81
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
+ K F++ +N+ H+MPTRY VS DFD+K L ++++ +K A +AL F +K +
Sbjct 82 KSTIKPFLKSINMNHVMPTRYVVS-DFDIKPLLQGIDMQEADGKKQALRALHLAFNDKLI 140
Query 127 NPVNE 131
N +E
Sbjct 141 NIQSE 145
> sce:YHR010W RPL27A, RPL27; L27A; K02901 large subunit ribosomal
protein L27e
Length=136
Score = 93.2 bits (230), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 53/125 (42%), Positives = 75/125 (60%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVS-TSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LK G+V VV+ GR AGKK V+V EG+K F + LVAGIE+ PL+V+K+ KV K
Sbjct 5 LKAGKVAVVVRGRYAGKKVVIVKPHDEGSKSHPFGHALVAGIERYPLKVTKKHGAKKVAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K FI+ VN HL+PTRYT+ + K++ + E + R+ AKK + F E+
Sbjct 65 RTKIKPFIKVVNYNHLLPTRYTLDVE-AFKSVVSTETFEQPSQREEAKKVVKKAFEERHQ 123
Query 127 NPVNE 131
N+
Sbjct 124 AGKNQ 128
> xla:380115 rpl27, MGC68687; ribosomal protein L27 (EC:3.6.5.3);
K02901 large subunit ribosomal protein L27e
Length=136
Score = 91.7 bits (226), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 47/125 (37%), Positives = 78/125 (62%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR AG+K V+V + +GT +R + + LVAGI++ P +V+ M ++ K
Sbjct 5 MKPGKVVLVLAGRYAGRKAVIVKNVDDGTSDRQYSHALVAGIDRYPRKVTATMGKKRIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K+F++ N HLMPTRY+V D KA+ + D A+++ A++ F E++
Sbjct 65 RSKIKSFVKVYNYNHLMPTRYSVDIPLD-KAVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> sce:YDR471W RPL27B; L27B; K02901 large subunit ribosomal protein
L27e
Length=136
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 52/125 (41%), Positives = 74/125 (59%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVVS-TSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LK G+V VV+ GR AGKK V+V EG+K F + LVAGIE+ P +V+K+ KV K
Sbjct 5 LKAGKVAVVVRGRYAGKKVVIVKPHDEGSKSHPFGHALVAGIERYPSKVTKKHGAKKVAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K FI+ VN HL+PTRYT+ + K++ + E + R+ AKK + F E+
Sbjct 65 RTKIKPFIKVVNYNHLLPTRYTLDVE-AFKSVVSTETFEQPSQREEAKKVVKKAFEERHQ 123
Query 127 NPVNE 131
N+
Sbjct 124 AGKNQ 128
> dre:325618 rpl27, fc93a08, wu:fc93a08, zgc:56171; ribosomal
protein L27; K02901 large subunit ribosomal protein L27e
Length=136
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR AG+K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVMVLAGRYAGRKAVIVKNIDDGTADRPYSHALVAGIDRYPRKVTATMGKKKIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + KAF++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKAFVKVFNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> ath:AT3G22230 60S ribosomal protein L27 (RPL27B); K02901 large
subunit ribosomal protein L27e
Length=135
Score = 89.4 bits (220), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 50/124 (40%), Positives = 75/124 (60%), Gaps = 3/124 (2%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LK + V++L GR AGKK V++ S +GT +R + +CLVAG++K P +V ++ S K K
Sbjct 5 LKQNKAVILLQGRYAGKKAVIIKSFDDGTSDRRYGHCLVAGLKKYPSKVIRKDSAKKTAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
+ R K FI+ VN +HLMPTRYT+ D D+K +A +L+ + +A K E+F
Sbjct 65 KSRVKCFIKLVNYQHLMPTRYTL--DVDLKEVATLDALKSKDKKVTALKEAKAKLEERFK 122
Query 127 NPVN 130
N
Sbjct 123 TGKN 126
> cel:C53H9.1 rpl-27; Ribosomal Protein, Large subunit family
member (rpl-27); K02901 large subunit ribosomal protein L27e
Length=136
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 69/106 (65%), Gaps = 1/106 (0%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL G+ AG+K VVV EG +R + + ++AGI++ PL+V+K M K++K
Sbjct 5 MKPGKVVLVLRGKYAGRKAVVVKQQDEGVSDRTYPHAIIAGIDRYPLKVTKDMGKKKIEK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKS 112
R + K F++ V+ HL+PTRY+V FD + EA S RK+
Sbjct 65 RNKLKPFLKVVSYTHLLPTRYSVDVAFDKTNINKEALKAPSKKRKA 110
> ath:AT2G32220 60S ribosomal protein L27 (RPL27A); K02901 large
subunit ribosomal protein L27e
Length=135
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 47/124 (37%), Positives = 74/124 (59%), Gaps = 3/124 (2%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+ V++L GR GKK V+V S +GT E+ + +CLVAG++K P +V ++ S K K
Sbjct 5 MKPGKAVILLQGRYTGKKAVIVKSFDDGTVEKKYGHCLVAGLKKYPSKVIRKDSAKKTAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
+ R K F + +N +H+MPTRYT+ D D+K + ++ + +A K F E+F
Sbjct 65 KSRVKCFFKVINYQHVMPTRYTL--DLDLKNVVSADAISSKDKKVTALKEAKAKFEERFK 122
Query 127 NPVN 130
N
Sbjct 123 TGKN 126
> mmu:19942 Rpl27, MGC103134; ribosomal protein L27 (EC:3.6.5.3);
K02901 large subunit ribosomal protein L27e
Length=136
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 46/125 (36%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR +G+K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGRYSGRKAVIVKNIDDGTSDRPYSHALVAGIDRYPRKVTAAMGKKKIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K+F++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKSFVKVYNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> mmu:100039431 Gm11518; 60S ribosomal protein L27; K02901 large
subunit ribosomal protein L27e
Length=136
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 46/125 (36%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR +G+K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGRYSGRKAVIVKNIDDGTSDRPYSHALVAGIDRYPRKVTAAMGKKKIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K+F++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKSFVKVYNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> hsa:6155 RPL27; ribosomal protein L27; K02901 large subunit
ribosomal protein L27e
Length=136
Score = 87.8 bits (216), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 46/125 (36%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR +G+K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGRYSGRKAVIVKNIDDGTSDRPYSHALVAGIDRYPRKVTAAMGKKKIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K+F++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKSFVKVYNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> ath:AT4G15000 60S ribosomal protein L27 (RPL27C); K02901 large
subunit ribosomal protein L27e
Length=131
Score = 87.4 bits (215), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 48/119 (40%), Positives = 74/119 (62%), Gaps = 3/119 (2%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
LK + V++L GR AGKK V++ S +G ++R + +CLVAG++K P +V ++ S K K
Sbjct 5 LKQNKAVILLQGRYAGKKAVIIKSFDDGNRDRPYGHCLVAGLKKYPSKVIRKDSAKKTAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKF 125
+ R K FI+ VN +HLMPTRYT+ D D+K +A +L+ + +A K E+F
Sbjct 65 KSRVKCFIKLVNYQHLMPTRYTL--DVDLKEVATLDALQSKDKKVAALKEAKAKLEERF 121
> mmu:622673 Gm6341, EG622673; predicted pseudogene 6341
Length=136
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 44/119 (36%), Positives = 74/119 (62%), Gaps = 2/119 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL G +G+K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGHYSGRKAVIVKNIDDGTSDRPYSHALVAGIDRYPRKVTAAMGKKKIAK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKF 125
R + K+F++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKSFVKVYNYNHLMPTRYSVDIPLD-KTVVKKDVFRDPALKRKARREAKVKFEERY 122
> mmu:100504769 60S ribosomal protein L27-like
Length=136
Score = 82.4 bits (202), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 75/125 (60%), Gaps = 2/125 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR + +K V+V + +GT +R + + LVAGI++ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGRYSRRKAVIVKNIDDGTLDRPYSHALVAGIDRYPRKVTAAMGKKKITK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKKALANIFYEKFM 126
R + K F++ N HLMPTRY+V D K + + D A+++ A++ F E++
Sbjct 65 RSKIKFFVKVYNYNHLMPTRYSVDIPLD-KTVVNKDVFRDPALKRKARREAKVKFEERYK 123
Query 127 NPVNE 131
N+
Sbjct 124 TGKNK 128
> mmu:223745 Gm4825, EG223745; predicted pseudogene 4825
Length=136
Score = 72.4 bits (176), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 39/109 (35%), Positives = 67/109 (61%), Gaps = 2/109 (1%)
Query 8 LKPGRVVVVLNGRMAGKKGVVV-STSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQK 66
+KPG+VV+VL GR +G K V++ + GT +R + LVAGI+ P +V+ M K+ K
Sbjct 5 MKPGKVVLVLAGRYSGHKAVIMKNIDNGTSDRPYSRVLVAGIDCYPQKVTAAMGKKKISK 64
Query 67 RMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKK 115
R + K+ ++ N HL+PTR++V+ D A+ + D A+++ A++
Sbjct 65 RSKFKSIVKVYNYNHLIPTRHSVAILLDKTAVNKDV-FRDPALKRKARQ 112
> dre:553755 atp5a1, zgc:154103; ATP synthase, H+ transporting,
mitochondrial F1 complex, alpha subunit 1, cardiac muscle;
K02132 F-type H+-transporting ATPase subunit alpha [EC:3.6.3.14]
Length=551
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 24/101 (23%), Positives = 45/101 (44%), Gaps = 18/101 (17%)
Query 7 PLKPGRVVVVLNGRMAGKKGVVVST-------SEGTKERHFCYCLVAGIEKAPLRVSKRM 59
P+ G+ +++ R GK + + T +EGT+E+ YC+ I + KR
Sbjct 200 PIGRGQRELIIGDRQTGKTAIAIDTIINQKRFNEGTEEKKKLYCIYVAIGQ------KRS 253
Query 60 STTKVQKRMRPKAFIRYVNVRHLMPTRYTVSNDFDVKALAP 100
+ ++ KR+ ++Y V T S+ ++ LAP
Sbjct 254 TVAQLVKRLTDADAMKYTIV-----VSATASDAAPLQYLAP 289
> ath:AT5G16486 hypothetical protein
Length=209
Score = 30.0 bits (66), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 11/22 (50%), Positives = 16/22 (72%), Gaps = 0/22 (0%)
Query 65 QKRMRPKAFIRYVNVRHLMPTR 86
+ R+ AFI +VN+RH +PTR
Sbjct 151 KGRIPKHAFISWVNIRHRLPTR 172
> ath:AT5G19400 SMG7; SMG7; K14409 protein SMG7
Length=1059
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 31/100 (31%), Positives = 48/100 (48%), Gaps = 23/100 (23%)
Query 20 RMAGK---KGVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTTKVQKRMRPKAF-IR 75
R+ GK KG +S + T LVAG EK + ++ M KAF IR
Sbjct 282 RLTGKGRGKGADISLKDAT--------LVAGPEKDKVTIANEML----------KAFSIR 323
Query 76 YVNVRHLMPTRYTVSNDFDVKALAPEASLEDSAMRKSAKK 115
+V++ ++ TR ++ FDV A + +SL + SAK+
Sbjct 324 FVHLNGILFTRTSLETFFDVLA-STSSSLREVISLGSAKE 362
> tgo:TGME49_055390 hypothetical protein
Length=2731
Score = 28.1 bits (61), Expect = 8.0, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Query 26 GVVVSTSEGTKERHFCYCLVAGIEKAPLRVSKRMSTT-KVQKRMR 69
G S EG +E HFC A +E+ P + T +VQ+R R
Sbjct 1838 GPTASADEGRRENHFC----AAVERPPESARRERGTRYEVQRRSR 1878
Lambda K H
0.319 0.130 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2105161088
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40