bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2703_orf1
Length=249
Score E
Sequences producing significant alignments: (Bits) Value
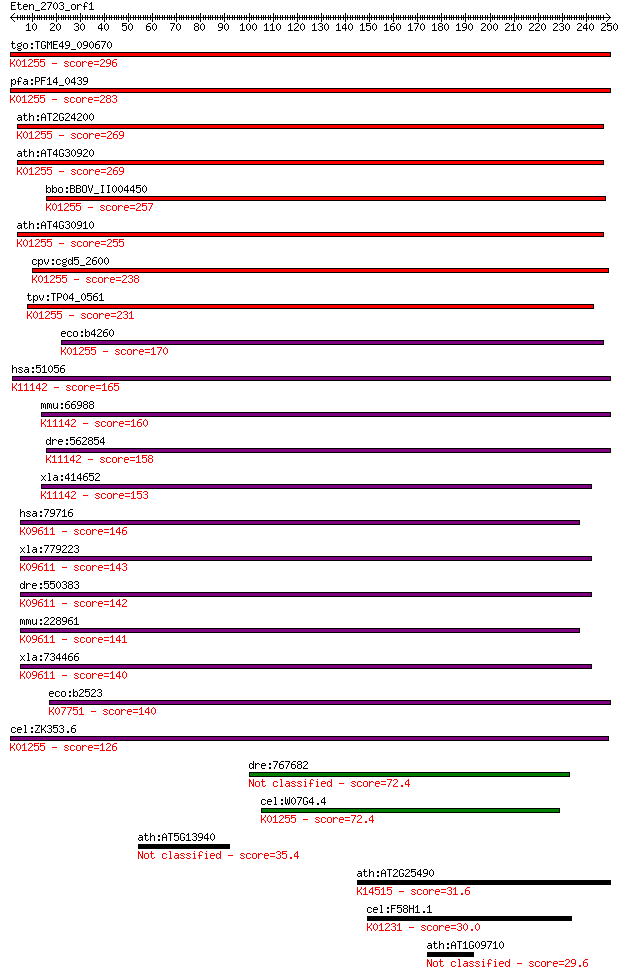
tgo:TGME49_090670 cytosol aminopeptidase (EC:3.4.11.1); K01255... 296 5e-80
pfa:PF14_0439 LAP; M17 leucyl aminopeptidase; K01255 leucyl am... 283 3e-76
ath:AT2G24200 cytosol aminopeptidase; K01255 leucyl aminopepti... 269 7e-72
ath:AT4G30920 cytosol aminopeptidase family protein; K01255 le... 269 7e-72
bbo:BBOV_II004450 18.m06372; leucine aminopeptidase; K01255 le... 257 2e-68
ath:AT4G30910 cytosol aminopeptidase family protein; K01255 le... 255 9e-68
cpv:cgd5_2600 leucine aminopeptidase; of possible plant or bac... 238 2e-62
tpv:TP04_0561 leucine aminopeptidase (EC:3.4.11.1); K01255 leu... 231 2e-60
eco:b4260 pepA, carP, ECK4253, JW4217, xerB; multifunctional a... 170 5e-42
hsa:51056 LAP3, LAP, LAPEP, PEPS; leucine aminopeptidase 3 (EC... 165 1e-40
mmu:66988 Lap3, 2410015L10Rik, AA410100, LAP-3, Lap, Lapep, Pe... 160 6e-39
dre:562854 lap3, im:7150907; leucine aminopeptidase 3 (EC:3.4.... 158 1e-38
xla:414652 lap3, MGC81140; leucine aminopeptidase 3; K11142 cy... 153 5e-37
hsa:79716 NPEPL1, FLJ11583, FLJ42065, bA261P9.2; aminopeptidas... 146 8e-35
xla:779223 lap-1, MGC154355; aminopeptidase-like 1; K09611 pro... 143 7e-34
dre:550383 npepl1, wu:fi27h09, wu:fj47h12, zgc:112459; aminope... 142 9e-34
mmu:228961 Npepl1, BC023239, C85514, mKIAA1974; aminopeptidase... 141 2e-33
xla:734466 npepl1, MGC115168; aminopeptidase-like 1; K09611 pr... 140 3e-33
eco:b2523 pepB, ECK2520, JW2507, yfhI; aminopeptidase B (EC:3.... 140 4e-33
cel:ZK353.6 lap-1; Leucine AminoPeptidase family member (lap-1... 126 7e-29
dre:767682 MGC152830, id:ibd2015, sb:cb283, wu:fb18d07; zgc:15... 72.4 1e-12
cel:W07G4.4 hypothetical protein; K01255 leucyl aminopeptidase... 72.4 1e-12
ath:AT5G13940 aminopeptidase 35.4 0.20
ath:AT2G25490 EBF1; EBF1 (EIN3-BINDING F BOX PROTEIN 1); prote... 31.6 3.1
cel:F58H1.1 aman-2; Alpha MANnosidase family member (aman-2); ... 30.0 8.3
ath:AT1G09710 DNA binding 29.6 9.4
> tgo:TGME49_090670 cytosol aminopeptidase (EC:3.4.11.1); K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=563
Score = 296 bits (758), Expect = 5e-80, Method: Compositional matrix adjust.
Identities = 165/253 (65%), Positives = 198/253 (78%), Gaps = 12/253 (4%)
Query 1 DGTEAVLQA---ARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQA 57
D EAV +A A+ VA GV+FA+ELVN+PANYC VTLA+AA +A E GLE ++L Q
Sbjct 226 DDVEAVNRAIARAKVVAPGVYFAQELVNAPANYCNPVTLARAAVEMAKEAGLEAEVLQQD 285
Query 58 EVEALGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYN 117
++E L MGCYL V KGS++PPQFIHLTY+ P R++AFVGKG+CFD+GGYN
Sbjct 286 DIEKLKMGCYLAVAKGSLFPPQFIHLTYKSSN--------PKRRVAFVGKGICFDAGGYN 337
Query 118 IKRAETSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVL 177
+K IELMKFDMGGAAAVLGAARALG L+P VEVHF+ AA ENM+S+++YRPGDV+
Sbjct 338 LKTGGAQIELMKFDMGGAAAVLGAARALGELKPENVEVHFLVAACENMISAKSYRPGDVI 397
Query 178 TASNGKTVEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFS 236
TASNGKTVEVGNTDAEGRLTLADALV+AEK ++ D I+DVATLTGA +VALG YAGL+S
Sbjct 398 TASNGKTVEVGNTDAEGRLTLADALVYAEKTVKADTIVDVATLTGAVIVALGYKYAGLWS 457
Query 237 PDDQLAEKILKCA 249
+ LA ILK A
Sbjct 458 NKECLASSILKSA 470
> pfa:PF14_0439 LAP; M17 leucyl aminopeptidase; K01255 leucyl
aminopeptidase [EC:3.4.11.1]
Length=605
Score = 283 bits (725), Expect = 3e-76, Method: Compositional matrix adjust.
Identities = 142/250 (56%), Positives = 181/250 (72%), Gaps = 8/250 (3%)
Query 1 DGTEAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVE 60
D + ++ AR G ++A +L+ +P+NYC V+L+ AA LA + LE KILG E+E
Sbjct 275 DTYKEEVEKARVYYFGTYYASQLIAAPSNYCNPVSLSNAAVELAQKLNLEYKILGVKELE 334
Query 61 ALGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKR 120
L MG YL V KGSMYP +FIHLTY+ KG +K+A VGKG+ FDSGGYN+K
Sbjct 335 ELKMGAYLSVGKGSMYPNKFIHLTYKSKGDVK-------KKIALVGKGITFDSGGYNLKA 387
Query 121 AETS-IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTA 179
A S I+LMKFDM G AAVLG A +G L+P VE+HF++A ENMVS +YRPGD++TA
Sbjct 388 APGSMIDLMKFDMSGCAAVLGCAYCVGTLKPENVEIHFLSAVCENMVSKNSYRPGDIITA 447
Query 180 SNGKTVEVGNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDD 239
SNGKT+EVGNTDAEGRLTLADALV+AEKL VD I+D+ATLTGA + +LG SYAG+F ++
Sbjct 448 SNGKTIEVGNTDAEGRLTLADALVYAEKLGVDYIVDIATLTGAMLYSLGTSYAGVFGNNE 507
Query 240 QLAEKILKCA 249
+L KIL +
Sbjct 508 ELINKILNSS 517
> ath:AT2G24200 cytosol aminopeptidase; K01255 leucyl aminopeptidase
[EC:3.4.11.1]
Length=520
Score = 269 bits (688), Expect = 7e-72, Method: Compositional matrix adjust.
Identities = 140/246 (56%), Positives = 173/246 (70%), Gaps = 9/246 (3%)
Query 4 EAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFG--LECKILGQAEVEA 61
E L+ A V+ GV F REL+NSPAN T LA+ A +A+ + IL + + +
Sbjct 189 EKKLKYAEDVSYGVIFGRELINSPANVLTPAVLAEEAAKVASTYSDVFTANILNEEQCKE 248
Query 62 LGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRA 121
L MG YL V S PP FIHL Y+P G+ KLA VGKGL FDSGGYNIK
Sbjct 249 LKMGSYLAVAAASANPPHFIHLVYKPPN------GSVKTKLALVGKGLTFDSGGYNIKTG 302
Query 122 E-TSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTAS 180
SIELMKFDMGG+AAVLGAA+A+G ++P GVEVHF+ AA ENM+S RPGDV+TAS
Sbjct 303 PGCSIELMKFDMGGSAAVLGAAKAIGEIKPPGVEVHFIVAACENMISGTGMRPGDVITAS 362
Query 181 NGKTVEVGNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQ 240
NGKT+EV NTDAEGRLTLADALV+A VD I+D+ATLTGACV+ALG S AG+++P D+
Sbjct 363 NGKTIEVNNTDAEGRLTLADALVYACNQGVDKIVDLATLTGACVIALGTSMAGIYTPSDE 422
Query 241 LAEKIL 246
LA++++
Sbjct 423 LAKEVI 428
> ath:AT4G30920 cytosol aminopeptidase family protein; K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=583
Score = 269 bits (688), Expect = 7e-72, Method: Compositional matrix adjust.
Identities = 140/246 (56%), Positives = 175/246 (71%), Gaps = 9/246 (3%)
Query 4 EAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFG--LECKILGQAEVEA 61
E L+ A HV+ GV F +ELVNSPAN T LA+ A +LA+ + + IL + + +
Sbjct 252 EKKLKYAEHVSYGVIFGKELVNSPANVLTPAVLAEEALNLASMYSDVMTANILNEEQCKE 311
Query 62 LGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRA 121
L MG YL V S PP FIHL Y+P + G KLA VGKGL FDSGGYNIK
Sbjct 312 LKMGSYLAVAAASANPPHFIHLIYKP------SSGPVKTKLALVGKGLTFDSGGYNIKTG 365
Query 122 ETS-IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTAS 180
IELMKFDMGG+AAVLGAA+A+G ++P GVEVHF+ AA ENM+S RPGDVLTAS
Sbjct 366 PGCLIELMKFDMGGSAAVLGAAKAIGQIKPPGVEVHFIVAACENMISGTGMRPGDVLTAS 425
Query 181 NGKTVEVGNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQ 240
NGKT+EV NTDAEGRLTLADALV+A VD ++D+ATLTGAC++ALG S AG+++P D+
Sbjct 426 NGKTIEVNNTDAEGRLTLADALVYACNQGVDKVVDLATLTGACIIALGTSMAGIYTPSDK 485
Query 241 LAEKIL 246
LA++++
Sbjct 486 LAKEVI 491
> bbo:BBOV_II004450 18.m06372; leucine aminopeptidase; K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=525
Score = 257 bits (657), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 127/232 (54%), Positives = 161/232 (69%), Gaps = 7/232 (3%)
Query 16 GVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGMGCYLGVCKGSM 75
G+H RELV +PANY T ++A ++ GLE +I+ + E ALGMG YL V +GS
Sbjct 210 GMHTTRELVTAPANYANTESIAGFLQNRLTGLGLEVRIIEEDECRALGMGAYLAVGQGSQ 269
Query 76 YPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFDMGGA 135
YPP+F+H YR + K+A VGKG+ FD+GG NIK A + IELMKFDMGG
Sbjct 270 YPPKFLHAIYRSSVSVK-------TKIALVGKGIMFDTGGLNIKSAASEIELMKFDMGGM 322
Query 136 AAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTDAEGR 195
+ V GAA + L+P GVEVHF++A ENM S AYRPGD++TASNGKT+EV NTDAEGR
Sbjct 323 STVFGAAETIAALKPHGVEVHFISATCENMAGSNAYRPGDIVTASNGKTIEVINTDAEGR 382
Query 196 LTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILK 247
LTLADALV+A+KL VD I+D+ATLTGAC++ALG Y G F D+ ++ K
Sbjct 383 LTLADALVYADKLGVDYIVDLATLTGACIIALGYQYGGYFVNDESFHQRFQK 434
> ath:AT4G30910 cytosol aminopeptidase family protein; K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=581
Score = 255 bits (652), Expect = 9e-68, Method: Compositional matrix adjust.
Identities = 134/246 (54%), Positives = 172/246 (69%), Gaps = 9/246 (3%)
Query 4 EAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFG--LECKILGQAEVEA 61
E L+ A HV+ GV F +ELVNSPAN + LA+ A +LA+ + + IL + + +
Sbjct 251 ENKLKYAEHVSYGVIFTKELVNSPANVLSPAVLAEEASNLASMYSNVMTANILKEEQCKE 310
Query 62 LGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRA 121
L MG YL V S PP FIHL Y+P + G KLA VGKGL FDSGGYNIK
Sbjct 311 LKMGSYLAVAAASANPPHFIHLIYKP------SSGPVKTKLALVGKGLTFDSGGYNIKIG 364
Query 122 -ETSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTAS 180
E IELMK D+GG+AAVLGAA+A+G ++P GVEVHF+ AA ENM+S RPGDV+TAS
Sbjct 365 PELIIELMKIDVGGSAAVLGAAKAIGEIKPPGVEVHFIVAACENMISGTGMRPGDVITAS 424
Query 181 NGKTVEVGNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQ 240
NGKT+EV +TD+EGRLTLADALV+A VD I+D+ATLTG +VALG S AG+++ D+
Sbjct 425 NGKTIEVNDTDSEGRLTLADALVYACNQGVDKIVDIATLTGEIIVALGPSMAGMYTASDE 484
Query 241 LAEKIL 246
LA++++
Sbjct 485 LAKEVI 490
> cpv:cgd5_2600 leucine aminopeptidase; of possible plant or bacterial
origin ; K01255 leucyl aminopeptidase [EC:3.4.11.1]
Length=550
Score = 238 bits (607), Expect = 2e-62, Method: Compositional matrix adjust.
Identities = 122/239 (51%), Positives = 161/239 (67%), Gaps = 9/239 (3%)
Query 10 ARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGMGCYLG 69
A+ V+ G+ FAR+L ++P NYC V +AK S+A GLE KIL E E L MG +L
Sbjct 232 AQSVSRGLLFARDLTSAPPNYCDPVNMAKEVISMAKSVGLEGKILQPKECEELKMGAFLA 291
Query 70 VCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMK 129
V +GS P QF+HLTY+PKG +++A VGKG+ D+GGYNIK I MK
Sbjct 292 VAQGSKSPAQFVHLTYKPKGEIK-------KRIALVGKGITMDTGGYNIKHQ--MIHFMK 342
Query 130 FDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGN 189
DMGGAAAV G A ++GL++P +EVHF++A +N+VS AY PG ++TASNGKT+EVGN
Sbjct 343 GDMGGAAAVFGTALSVGLIKPENIEVHFISAICDNLVSRDAYLPGCIITASNGKTIEVGN 402
Query 190 TDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILKC 248
TDAEGRLTLADALV+A L+V+ ++D+ATLTGA YA + +D+L + I C
Sbjct 403 TDAEGRLTLADALVYACNLKVETVIDLATLTGANYRLFEGRYASVLGNNDELFQMIQTC 461
> tpv:TP04_0561 leucine aminopeptidase (EC:3.4.11.1); K01255 leucyl
aminopeptidase [EC:3.4.11.1]
Length=541
Score = 231 bits (590), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 114/235 (48%), Positives = 154/235 (65%), Gaps = 7/235 (2%)
Query 8 QAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGMGCY 67
Q + + + A++L ++P NYC TVT+++ + GL+ K+L + + L M CY
Sbjct 201 QRVKAFSESMDMAKQLTSAPPNYCNTVTVSEFFKKKLTSLGLKVKVLEYDDCKKLNMFCY 260
Query 68 LGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIEL 127
L V +GS +PP+F+H Y+P+G ++LAFVGKGL FDSGGYNIK + + I
Sbjct 261 LSVAQGSKFPPKFLHAVYKPEGEVK-------KRLAFVGKGLMFDSGGYNIKSSSSMIHY 313
Query 128 MKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEV 187
MK DM G V AA AL L+P VE+HFV ENM+ + +YRPGDV+TASNGKTVEV
Sbjct 314 MKLDMAGFGTVFSAAYALAKLKPNNVELHFVGGLCENMLDANSYRPGDVVTASNGKTVEV 373
Query 188 GNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLA 242
NTDAEGR+TLAD+L +A +L D ++D ATLTGA +VALG A +S D+ LA
Sbjct 374 LNTDAEGRVTLADSLYYAAQLNPDYLVDCATLTGAAMVALGVKCAAFYSNDEDLA 428
> eco:b4260 pepA, carP, ECK4253, JW4217, xerB; multifunctional
aminopeptidase A: a cyteinylglycinase, transcription regulator
and site-specific recombination factor (EC:3.4.11.1); K01255
leucyl aminopeptidase [EC:3.4.11.1]
Length=503
Score = 170 bits (430), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 99/227 (43%), Positives = 135/227 (59%), Gaps = 10/227 (4%)
Query 22 ELVNSPANYCTTVTLAKAAESLAAEF--GLECKILGQAEVEALGMGCYLGVCKGSMYPPQ 79
+L N P N C LA A LA + + +++G+ +++ LGM YL V +GS
Sbjct 189 DLGNMPPNICNAAYLASQARQLADSYSKNVITRVIGEQQMKELGMHSYLAVGQGSQNESL 248
Query 80 FIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFDMGGAAAVL 139
+ Y+ + D P + VGKGL FDSGG +IK +E ++ MK+DM GAAAV
Sbjct 249 MSVIEYKGNASEDARP------IVLVGKGLTFDSGGISIKPSE-GMDEMKYDMCGAAAVY 301
Query 140 GAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTDAEGRLTLA 199
G R + LQ + V V A ENM +AYRPGDVLT +G+TVEV NTDAEGRL L
Sbjct 302 GVMRMVAELQ-LPINVIGVLAGCENMPGGRAYRPGDVLTTMSGQTVEVLNTDAEGRLVLC 360
Query 200 DALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKIL 246
D L + E+ E +A++DVATLTGACV+ALG GL + + LA +++
Sbjct 361 DVLTYVERFEPEAVIDVATLTGACVIALGHHITGLMANHNPLAHELI 407
> hsa:51056 LAP3, LAP, LAPEP, PEPS; leucine aminopeptidase 3 (EC:3.4.11.5
3.4.11.1); K11142 cytosol aminopeptidase [EC:3.4.11.1
3.4.11.5]
Length=519
Score = 165 bits (418), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 101/250 (40%), Positives = 140/250 (56%), Gaps = 10/250 (4%)
Query 2 GTEAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAES--LAAEFGLECKILGQAEV 59
G + Q A+G + AR+L+ +PAN T A+ E +A E I ++ +
Sbjct 181 GDQEAWQKGVLFASGQNLARQLMETPANEMTPTRFAEIIEKNLKSASSKTEVHIRPKSWI 240
Query 60 EALGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIK 119
E MG +L V KGS PP F+ + Y+ G+P A L FVGKG+ FDSGG +IK
Sbjct 241 EEQAMGSFLSVAKGSDEPPVFLEIHYK------GSPNANEPPLVFVGKGITFDSGGISIK 294
Query 120 RAETSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTA 179
A +++LM+ DMGGAA + A + L + + +A ENM S +A +PGDV+ A
Sbjct 295 -ASANMDLMRADMGGAATICSAIVSAAKLN-LPINIIGLAPLCENMPSGKANKPGDVVRA 352
Query 180 SNGKTVEVGNTDAEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDD 239
NGKT++V NTDAEGRL LADAL +A IL+ ATLTGA VALG G+F+
Sbjct 353 KNGKTIQVDNTDAEGRLILADALCYAHTFNPKVILNAATLTGAMDVALGSGATGVFTNSS 412
Query 240 QLAEKILKCA 249
L K+ + +
Sbjct 413 WLWNKLFEAS 422
> mmu:66988 Lap3, 2410015L10Rik, AA410100, LAP-3, Lap, Lapep,
Pep-7, Pep-S, Pep7, Peps; leucine aminopeptidase 3 (EC:3.4.11.5
3.4.11.1); K11142 cytosol aminopeptidase [EC:3.4.11.1 3.4.11.5]
Length=519
Score = 160 bits (404), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 98/238 (41%), Positives = 135/238 (56%), Gaps = 10/238 (4%)
Query 14 AAGVHFARELVNSPANYCTTVTLAKAAES--LAAEFGLECKILGQAEVEALGMGCYLGVC 71
A+G + AR L+ SPAN T A+ E +A + I ++ +E MG +L V
Sbjct 193 ASGQNLARHLMESPANEMTPTRFAEIIEKNLKSASSKTKVHIRPKSWIEEQEMGSFLSVA 252
Query 72 KGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFD 131
KGS PP F+ + Y G+P A L FVGKG+ FDSGG +IK A +++LM+ D
Sbjct 253 KGSEEPPVFLEIHYM------GSPNATEAPLVFVGKGITFDSGGISIK-ASANMDLMRAD 305
Query 132 MGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTD 191
MGGAA + A + L + + +A ENM S +A +PGDV+ A NGKT++V NTD
Sbjct 306 MGGAATICSAIVSAAKLN-LPINIIGLAPLCENMPSGKANKPGDVVRARNGKTIQVDNTD 364
Query 192 AEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILKCA 249
AEGRL LADAL +A I++ ATLTGA VALG G+F+ L K+ + +
Sbjct 365 AEGRLILADALCYAHTFNPKVIINAATLTGAMDVALGSGATGVFTNSSWLWNKLFEAS 422
> dre:562854 lap3, im:7150907; leucine aminopeptidase 3 (EC:3.4.11.1);
K11142 cytosol aminopeptidase [EC:3.4.11.1 3.4.11.5]
Length=517
Score = 158 bits (400), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 99/237 (41%), Positives = 138/237 (58%), Gaps = 12/237 (5%)
Query 16 GVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKI--LGQAEVEALGMGCYLGVCKG 73
G + AR+L+ +PAN+ T A E + F + Q+ +E MG +L V KG
Sbjct 193 GQNLARQLMEAPANHVTPTVFANTIEQKLSLFADRVIVHKRPQSWIEGEQMGAFLSVSKG 252
Query 74 SMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFDMG 133
S PP F+ L Y KG++D A L VGKG+ FDSGG ++K + ++ M+ DMG
Sbjct 253 SEEPPVFLELHY--KGSSDSAQPP----LVLVGKGITFDSGGISLK-PSSGMDAMRADMG 305
Query 134 GAAAVLGA-ARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTDA 192
GAA V A A L P + + +A ENM S +A +PGDV+ A NGKT+++ NTDA
Sbjct 306 GAATVCSAIVTAAALKIP--INIIGLAPLCENMPSGKANKPGDVVRAKNGKTIQIDNTDA 363
Query 193 EGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILKCA 249
EGRL LADAL +A + AI++ ATLTGA VALG + G+F+ D L E++ K +
Sbjct 364 EGRLILADALCYAHSFKPRAIINAATLTGAMDVALGSAATGVFTNSDWLWERLHKAS 420
> xla:414652 lap3, MGC81140; leucine aminopeptidase 3; K11142
cytosol aminopeptidase [EC:3.4.11.1 3.4.11.5]
Length=495
Score = 153 bits (387), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 95/230 (41%), Positives = 131/230 (56%), Gaps = 10/230 (4%)
Query 14 AAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAE--VEALGMGCYLGVC 71
A G + AR L+ +PANY T A+ E A K+ +++ +E MG +L V
Sbjct 167 AEGQNLARHLMEAPANYITPTKFAEIFEQKLASMDSSVKVFTRSKPWIEEQQMGAFLSVA 226
Query 72 KGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFD 131
KGS PP F+ + Y G+ A L FVGKG+ FDSGG ++K + + ++ M+ D
Sbjct 227 KGSEEPPIFLEIHY------SGSSDASQPPLVFVGKGVTFDSGGISLKPS-SGMDAMRAD 279
Query 132 MGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTD 191
MGGAA + A L+ + + +A ENM S A +PGDV+ A NGKT++V NTD
Sbjct 280 MGGAATICSAIMIAAKLK-LPINIIGLAPLCENMPSGGANKPGDVVKAKNGKTIQVDNTD 338
Query 192 AEGRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQL 241
AEGRL LADAL +A AI++ ATLTGA VALG + AG+F+ L
Sbjct 339 AEGRLILADALCYAHSFNPRAIVNAATLTGAMDVALGSAAAGVFTNSSWL 388
> hsa:79716 NPEPL1, FLJ11583, FLJ42065, bA261P9.2; aminopeptidase-like
1; K09611 probable aminopeptidase NPEPL1 [EC:3.4.11.-]
Length=523
Score = 146 bits (368), Expect = 8e-35, Method: Compositional matrix adjust.
Identities = 94/233 (40%), Positives = 133/233 (57%), Gaps = 11/233 (4%)
Query 5 AVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGM 64
+ LQ + GV A +V++P N T T + + E G+ I+ E++ G
Sbjct 166 STLQCLANATDGVRLAARIVDTPCNEMNTDTFLEEINKVGKELGIIPTIIRDEELKTRGF 225
Query 65 GCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETS 124
G GV K +++PP L++ P GAT + +A+VGKG+ +D+GG +IK +T+
Sbjct 226 GGIYGVGKAALHPPALAVLSHTPDGAT--------QTIAWVGKGIVYDTGGLSIK-GKTT 276
Query 125 IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKT 184
+ MK D GGAAAVLGA RA + Q +H V AEN V A RP D+ +GKT
Sbjct 277 MPGMKRDCGGAAAVLGAFRA-AIKQGFKDNLHAVFCLAENSVGPNATRPDDIHLLYSGKT 335
Query 185 VEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFS 236
VE+ NTDAEGRL LAD + +A K L D ILD+ATLTGA +A G+ +A + +
Sbjct 336 VEINNTDAEGRLVLADGVSYACKDLGADIILDMATLTGAQGIATGKYHAAVLT 388
> xla:779223 lap-1, MGC154355; aminopeptidase-like 1; K09611 probable
aminopeptidase NPEPL1 [EC:3.4.11.-]
Length=521
Score = 143 bits (360), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 90/238 (37%), Positives = 138/238 (57%), Gaps = 11/238 (4%)
Query 5 AVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGM 64
+ L+ GV A +V++P + T + +++A + G+ I+ E++ G
Sbjct 166 STLKCLESATEGVRLAARIVDTPCSEMNTDHFIEEIKAVALDLGITPAIIRDVELKQRGF 225
Query 65 GCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETS 124
G GV K + +PP + L++ P+GAT + +A+VGKG+ +D+GG +IK +T+
Sbjct 226 GGIYGVGKAAEHPPALVILSHTPEGAT--------QTIAWVGKGIVYDTGGLSIK-GKTT 276
Query 125 IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKT 184
+ MK D GGAAAVLGA +A + Q +H + AEN V +A RP D+ +GKT
Sbjct 277 MPGMKRDCGGAAAVLGAFKA-AVKQGFKDNLHALFCLAENSVGPKATRPDDIHVLYSGKT 335
Query 185 VEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFSPDDQL 241
VE+ NTDAEGRL LAD + +A K L D ILD+ATLTGA +A G+ +A + + D+
Sbjct 336 VEINNTDAEGRLVLADGVSYACKDLGADIILDMATLTGAQSIATGKYHAAVLTNSDEW 393
> dre:550383 npepl1, wu:fi27h09, wu:fj47h12, zgc:112459; aminopeptidase-like
1; K09611 probable aminopeptidase NPEPL1 [EC:3.4.11.-]
Length=525
Score = 142 bits (359), Expect = 9e-34, Method: Compositional matrix adjust.
Identities = 90/238 (37%), Positives = 137/238 (57%), Gaps = 11/238 (4%)
Query 5 AVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGM 64
+ L+ + A GV A +V++P + T + +++ G+ I+ E++ G
Sbjct 166 STLECLANAADGVRLAARIVDTPCSEMNTDDFLEEIKTVGNALGITPTIIRGEELKQKGF 225
Query 65 GCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETS 124
G GV K ++ PP L++ P GAT + +A+VGKG+ +D+GG +IK +T+
Sbjct 226 GGIYGVGKAALNPPALAVLSHTPSGAT--------QTIAWVGKGIVYDTGGLSIK-GKTT 276
Query 125 IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKT 184
+ MK D GGAAA+LGA +A + Q +H V AEN V A RP D+ T +GKT
Sbjct 277 MPGMKRDCGGAAAILGAFKAT-IKQGFKDNLHAVFCLAENSVGPAATRPDDIHTLYSGKT 335
Query 185 VEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFSPDDQL 241
VE+ NTDAEGRL L+D +V+A K L D ILD+ATLTGA ++ G+ +A + + +Q
Sbjct 336 VEINNTDAEGRLVLSDGVVYASKDLSADIILDMATLTGAQGISTGKHHAAVMTNSEQW 393
> mmu:228961 Npepl1, BC023239, C85514, mKIAA1974; aminopeptidase-like
1; K09611 probable aminopeptidase NPEPL1 [EC:3.4.11.-]
Length=524
Score = 141 bits (356), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 91/233 (39%), Positives = 132/233 (56%), Gaps = 11/233 (4%)
Query 5 AVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGM 64
+ LQ + GV A +V++P N T + + E G+ I+ +++ G
Sbjct 166 STLQCLTNATEGVRLAARIVDTPCNEMNTDIFLEEIIQVGKELGITPTIIRDEQLKTKGF 225
Query 65 GCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETS 124
G GV K +++PP L++ P GAT + +A+VGKG+ +D+GG +IK +T+
Sbjct 226 GGIYGVGKAALHPPALAILSHTPDGAT--------QTIAWVGKGIVYDTGGLSIK-GKTT 276
Query 125 IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKT 184
+ MK D GGAAAVLGA RA + Q +H V AEN V A RP D+ +GKT
Sbjct 277 MPGMKRDCGGAAAVLGAFRA-AIKQGFKDNLHAVFCLAENAVGPNATRPDDIHLLYSGKT 335
Query 185 VEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFS 236
VE+ NTDAEGRL LAD + +A K L D I+D+ATLTGA +A G+ +A + +
Sbjct 336 VEINNTDAEGRLVLADGVSYACKDLGADIIVDMATLTGAQGIATGKYHAAVLT 388
> xla:734466 npepl1, MGC115168; aminopeptidase-like 1; K09611
probable aminopeptidase NPEPL1 [EC:3.4.11.-]
Length=521
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 89/238 (37%), Positives = 138/238 (57%), Gaps = 11/238 (4%)
Query 5 AVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGM 64
+ L+ GV A +V++P + T + +++A + G+ I+ E++ G
Sbjct 166 STLKCLESATEGVRLAARIVDTPCSEMNTDHFVEEIKAVAQDLGITPVIIRDEELKQRGF 225
Query 65 GCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETS 124
G GV K + +PP + L++ P+GAT + +A+VGKG+ +D+GG +IK +T+
Sbjct 226 GGIYGVGKAAEHPPALVILSHTPEGAT--------QTIAWVGKGIVYDTGGLSIK-GKTT 276
Query 125 IELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKT 184
+ MK D GGAAAVLGA +A + Q +H + AEN V +A RP D+ +GKT
Sbjct 277 MPGMKRDCGGAAAVLGAFKA-AVKQGFKDNLHALFCLAENSVGPKATRPDDIHVLYSGKT 335
Query 185 VEVGNTDAEGRLTLADALVFAEK-LEVDAILDVATLTGACVVALGESYAGLFSPDDQL 241
VE+ NTDAEGRL LAD + +A K L D ILD+ATLTGA +A G+ +A + + ++
Sbjct 336 VEINNTDAEGRLVLADGVSYACKDLGADIILDMATLTGAQGIATGKHHAAVLTNSEEW 393
> eco:b2523 pepB, ECK2520, JW2507, yfhI; aminopeptidase B (EC:3.4.11.23);
K07751 PepB aminopeptidase [EC:3.4.11.23]
Length=427
Score = 140 bits (353), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 94/236 (39%), Positives = 128/236 (54%), Gaps = 10/236 (4%)
Query 17 VHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVEALGMGCYLG---VCKG 73
+ + R+ +N+PA LA+ A L + + + E L Y+G V +G
Sbjct 107 IDWVRDTINAPAEELGPSQLAQRAVDLISNVAGDRVTYRITKGEDLREQGYMGLHTVGRG 166
Query 74 SMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKRAETSIELMKFDMG 133
S P + L Y P G + AP+ VGKG+ FDSGGY+IK+ ++ MK DMG
Sbjct 167 SERSPVLLALDYNPTGDKE----APVYA-CLVGKGITFDSGGYSIKQT-AFMDSMKSDMG 220
Query 134 GAAAVLGAARALGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTDAE 193
GAA V GA A + + V A+N++S A++ GD++T NGK VEV NTDAE
Sbjct 221 GAATVTGAL-AFAITRGLNKRVKLFLCCADNLISGNAFKLGDIITYRNGKKVEVMNTDAE 279
Query 194 GRLTLADALVFAEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILKCA 249
GRL LAD L+ A + + I+D ATLTGA ALG Y LFS DD LA ++L A
Sbjct 280 GRLVLADGLIDASAQKPEMIIDAATLTGAAKTALGNDYHALFSFDDALAGRLLASA 335
> cel:ZK353.6 lap-1; Leucine AminoPeptidase family member (lap-1);
K01255 leucyl aminopeptidase [EC:3.4.11.1]
Length=491
Score = 126 bits (317), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 91/252 (36%), Positives = 129/252 (51%), Gaps = 28/252 (11%)
Query 1 DGTEAVLQAARHVAAGVHFARELVNSPANYCTTVTLAKAAESLAAEFGLECKILGQAEVE 60
D +E+V + AR L+++PAN TT L A + G + ++ E+
Sbjct 165 DLSESVRETAR-----------LIDTPANILTTDALVDEAVKVGNATGSKITVIRGEELL 213
Query 61 ALGMGCYLGVCKGSMYPPQFIHLTYRPKGATDGAPGAPLRKLAFVGKGLCFDSGGYNIKR 120
G G V K PP F+ L++ G+T+ +A VGKG+ +D+GG IK
Sbjct 214 KAGFGGIYHVGKAGPTPPAFVVLSHEVPGSTE--------HIALVGKGVVYDTGGLQIK- 264
Query 121 AETSIELMKFDMGGAAAVLGAARAL---GLLQPAGVEVHFVAAAAENMVSSQAYRPGDVL 177
+T + MK DMGGAA +L A AL G Q +H EN VS A +P D++
Sbjct 265 TKTGMPNMKRDMGGAAGMLEAYSALVKHGFSQ----TLHACLCIVENNVSPIANKPDDII 320
Query 178 TASNGKTVEVGNTDAEGRLTLADALVFA-EKLEVDAILDVATLTGACVVALGESYAGLFS 236
+GKTVE+ NTDAEGRL LAD + +A E L+ I D+ATLTGA G + +
Sbjct 321 KMLSGKTVEINNTDAEGRLILADGVFYAKETLKATTIFDMATLTGAQAWLSGRLHGAAMT 380
Query 237 PDDQLAEKILKC 248
D+QL +I+K
Sbjct 381 NDEQLENEIIKA 392
> dre:767682 MGC152830, id:ibd2015, sb:cb283, wu:fb18d07; zgc:152830
Length=512
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/137 (34%), Positives = 74/137 (54%), Gaps = 5/137 (3%)
Query 100 RKLAFVGKGLCFDSGGYNIKRAETSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVA 159
+ L VGKG+ +D+GG +IK A + M D G+AAV G + L L+P ++V
Sbjct 255 QTLMLVGKGITYDTGGADIK-AGGIMAGMHGDKCGSAAVAGFFQILAKLKPKHLKVVGAM 313
Query 160 AAAENMVSSQAYRPGDVLTASNGKTVEVGNTDAEGRLTLADALVFAEKLEVDAI----LD 215
A N V S Y +++ + G+ + VGNTDAEGR+ + D L ++ + +
Sbjct 314 AMVRNSVGSDCYVADELVVSRAGRRIRVGNTDAEGRMVMVDLLCEMKEQALQEVSPHLFT 373
Query 216 VATLTGACVVALGESYA 232
+ATLTG + A+G Y+
Sbjct 374 IATLTGHAIRAMGPKYS 390
> cel:W07G4.4 hypothetical protein; K01255 leucyl aminopeptidase
[EC:3.4.11.1]
Length=522
Score = 72.4 bits (176), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 45/128 (35%), Positives = 73/128 (57%), Gaps = 5/128 (3%)
Query 105 VGKGLCFDSGGYNIKRAETSIELMKFDMGGAAAVLGAARALGLLQPAGVEVHFVAAAAEN 164
VGKG+ D+GG ++K + + D G+A V G +A+ +L+P ++ N
Sbjct 269 VGKGVTIDTGGCDLKTGGHMFGMCR-DKYGSAVVGGFFKAIDVLKPKNIKAVGYMCMVRN 327
Query 165 MVSSQAYRPGDVLTASNGKTVEVGNTDAEGRLTLADALVFAEKLEVDA----ILDVATLT 220
+ S AY +V+T+ +GK + + NTDAEGRLT+ D L A++ ++A + VATLT
Sbjct 328 SIGSHAYTCDEVITSRSGKRIHIYNTDAEGRLTMLDPLTLAKEEALNAKNPHLFTVATLT 387
Query 221 GACVVALG 228
G V++ G
Sbjct 388 GHEVLSYG 395
> ath:AT5G13940 aminopeptidase
Length=809
Score = 35.4 bits (80), Expect = 0.20, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 54 LGQAEVEALGMGCYLGVCKGSMYPPQFIHLTYRPKGAT 91
L + + L MG YL V S P FIHL Y+P +
Sbjct 77 LNEERCKELKMGTYLSVAAASANPHHFIHLVYKPASGS 114
> ath:AT2G25490 EBF1; EBF1 (EIN3-BINDING F BOX PROTEIN 1); protein
binding / ubiquitin-protein ligase; K14515 EIN3-binding
F-box protein
Length=628
Score = 31.6 bits (70), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 49/105 (46%), Gaps = 12/105 (11%)
Query 145 LGLLQPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGNTDAEGRLTLADALVF 204
L L+Q + V+++F + N+ R +TA NG T+EV N D +T A +
Sbjct 486 LHLIQSSLVKINF--SGCSNLTD----RVISAITARNGWTLEVLNIDGCSNITDASLVSI 539
Query 205 AEKLEVDAILDVATLTGACVVALGESYAGLFSPDDQLAEKILKCA 249
A ++ + LD++ A+ +S + D+L +IL A
Sbjct 540 AANCQILSDLDISK------CAISDSGIQALASSDKLKLQILSVA 578
> cel:F58H1.1 aman-2; Alpha MANnosidase family member (aman-2);
K01231 alpha-mannosidase II [EC:3.2.1.114]
Length=1145
Score = 30.0 bits (66), Expect = 8.3, Method: Composition-based stats.
Identities = 24/88 (27%), Positives = 36/88 (40%), Gaps = 5/88 (5%)
Query 149 QPAGVEVHFVAAAAENMVSSQAYRPGDVLTASNGKTVEVGN---TDAEGRLTLADALVFA 205
Q ++H V + S Y + TA+ K V + D RLTL D A
Sbjct 1040 QELNCDLHLVTF--RTLASPTTYEANERSTAAEKKAAMVMHRVVPDCRSRLTLPDTSCLA 1097
Query 206 EKLEVDAILDVATLTGACVVALGESYAG 233
LE++ + ++TL A +L Y G
Sbjct 1098 TGLEIEPLKLISTLKSAKKTSLTNLYEG 1125
> ath:AT1G09710 DNA binding
Length=610
Score = 29.6 bits (65), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 13/19 (68%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 174 GDVLTASNGKTVEVGNTDA 192
GDVLTA++G+ VE G TDA
Sbjct 367 GDVLTAASGRKVEPGKTDA 385
Lambda K H
0.318 0.134 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8892779092
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40