bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2657_orf1
Length=252
Score E
Sequences producing significant alignments: (Bits) Value
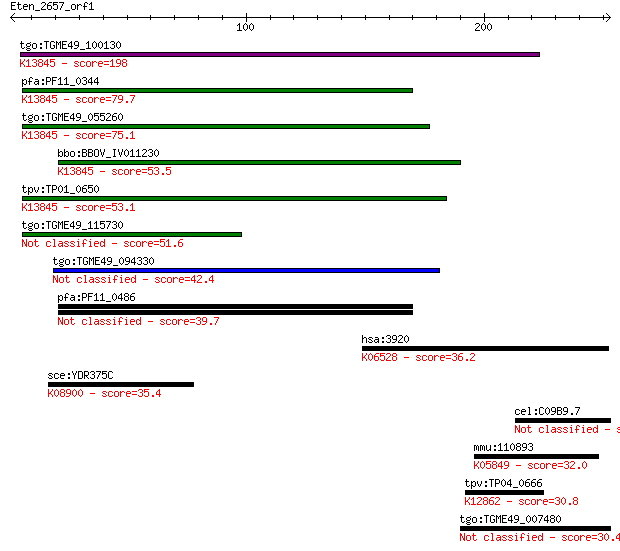
tgo:TGME49_100130 apical membrane antigen, putative ; K13845 a... 198 1e-50
pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane ... 79.7 9e-15
tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845... 75.1 2e-13
bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845... 53.5 7e-07
tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoi... 53.1 9e-07
tgo:TGME49_115730 apical membrane antigen, putative 51.6 3e-06
tgo:TGME49_094330 EGF-like domain-containing protein (EC:2.7.1... 42.4 0.002
pfa:PF11_0486 MAEBL, putative 39.7 0.012
hsa:3920 LAMP2, CD107b, LAMPB, LGP110; lysosomal-associated me... 36.2 0.12
sce:YDR375C BCS1; Bcs1p; K08900 mitochondrial chaperone BCS1 35.4 0.22
cel:C09B9.7 hypothetical protein 35.0 0.25
mmu:110893 Slc8a3, AW742262, MGC90626, Ncx3; solute carrier fa... 32.0 2.0
tpv:TP04_0666 hypothetical protein; K12862 pleiotropic regulat... 30.8 4.8
tgo:TGME49_007480 hypothetical protein 30.4 6.8
> tgo:TGME49_100130 apical membrane antigen, putative ; K13845
apical merozoite antigen 1
Length=493
Score = 198 bits (504), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 100/226 (44%), Positives = 136/226 (60%), Gaps = 10/226 (4%)
Query 5 GKYCSVGDASPVLTWYCFYPEKTT-RPVSYNSPYVREDHATACPEKAILGAHFGTWDGTT 63
G YC GD+ P LTWYCF+PEK+ + + + S Y R DHA+ACPE + H+G W+G +
Sbjct 236 GTYCKRGDSGPNLTWYCFHPEKSIEKNLVWGSAYARLDHASACPEHGLKNVHWGQWNGRS 295
Query 64 CQRMKAAKQITVPNPTECGKAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATMWPVGA 123
CQ+M K+I+V + TEC +F S SDNPTQY + V + P G+
Sbjct 296 CQKMAVRKRISVGSATECAMELFNNSPSDNPTQYVGDEGRGWDKIL--DDVVGVLIPAGS 353
Query 124 FSKDEPRTQGVGTNYANWY---TNGTCEMYDMVPTCFTLAPNQFSFTSLGSADPSTAELP 180
KD+P T+GVG N+AN+Y + CEMYD VP C T AP Q++F SLG +P A+LP
Sbjct 354 TKKDQPHTRGVGINWANFYKKPSGSYCEMYDGVPNCLTSAPEQYAFVSLGDPNPDNAQLP 413
Query 181 PCTEASEGWEIYGYCECGDGH----STPWKCENGQWIGGSDDCNCS 222
PC+ A+EG I +C C +G S+ KCE+G+W+ G C CS
Sbjct 414 PCSSATEGVVIPSHCSCPEGETSGSSSGVKCEDGKWVEGHVSCTCS 459
> pfa:PF11_0344 AMA1, AMA-1, Pf83, RMA-1, RMA1; apical membrane
antigen 1, AMA1; K13845 apical merozoite antigen 1
Length=622
Score = 79.7 bits (195), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 51/172 (29%), Positives = 80/172 (46%), Gaps = 9/172 (5%)
Query 6 KYCSVGDASPVLTWYCFYPEKTT--RPVSYNSPYVREDHATACPEKAILGAHFGTWDGTT 63
+YC+ D S + +CF P K + +Y S V ++ CP K + A FG W
Sbjct 261 RYCN-KDESKRNSMFCFRPAKDISFQNYTYLSKNVVDNWEKVCPRKNLQNAKFGLWVDGN 319
Query 64 CQRMKAAKQITVPNPTECGKAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATM----- 118
C+ + + + EC K VF++S+SD P QY + T N A+M
Sbjct 320 CEDIPHVNEFPAIDLFECNKLVFELSASDQPKQYEQHLTDYEKIKEGFKNKNASMIKSAF 379
Query 119 WPVGAFSKDEPRTQGVGTNYANWYTN-GTCEMYDMVPTCFTLAPNQFSFTSL 169
P GAF D ++ G G N+ N+ T CE++++ PTC + + T+L
Sbjct 380 LPTGAFKADRYKSHGKGYNWGNYNTETQKCEIFNVKPTCLINNSSYIATTAL 431
> tgo:TGME49_055260 apical membrane antigen 1, putative ; K13845
apical merozoite antigen 1
Length=569
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 59/186 (31%), Positives = 81/186 (43%), Gaps = 27/186 (14%)
Query 6 KYCSVGDASPVLTWYCFYPEKTT---RPVSYNSPYVRED---HATACPEKAILGAHFGTW 59
KYCSV P LTWYCF P K+ + Y S YV E+ + CP +A+ G FG W
Sbjct 240 KYCSVKGEPPDLTWYCFKPRKSVTENHHLIYGSAYVGENPDAFISKCPNQALRGYRFGVW 299
Query 60 DGTTCQRMKAAKQIT---VPNPTECGKAVFKVS--SSDNPTQYTKPPTTEASSSTSSSNA 114
C V + +C F+ +SD P +T P T++AS +
Sbjct 300 KKGRCLDYTELTDTVIERVESKAQCWVKTFENDGVASDQP--HTYPLTSQASWN------ 351
Query 115 VATMWPVGAFSKDEPRTQGVGTNYANWYTN----GTCEMYDMVPTCFTLAPNQFSFTSLG 170
WP+ D+P + GVG NY +Y + G C + D VP C S+T+ G
Sbjct 352 --DWWPL--HQSDQPHSGGVGRNYGFYYVDTTGEGKCALSDQVPDCLVSDSAAVSYTAAG 407
Query 171 SADPST 176
S T
Sbjct 408 SLSEET 413
> bbo:BBOV_IV011230 23.m06405; apical membrane antigen 1; K13845
apical merozoite antigen 1
Length=605
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 45/179 (25%), Positives = 70/179 (39%), Gaps = 10/179 (5%)
Query 21 CFYPEKTTRP--VSYNSPYVREDHATACPEKAILGAHFGTWDGTTCQRMKAAKQITVPNP 78
C P K+ + Y S V D CP + A FG W G +C + A Q +
Sbjct 275 CMKPYKSAEDAHLYYGSAKVDPDWEENCPMHPVRDAIFGKWSGGSCVAIAPAFQEYANST 334
Query 79 TECGKAVFKVSSSDNPTQYTKPPTTEASSSTS-------SSNAVATMWPVGAFSKDEPRT 131
+C +F S++D + E TS S A A P+ + +
Sbjct 335 EDCAAILFDNSATDLDIEVVNEEFNELKELTSGLKRLNLSKVANAIFSPLSNVAGTSRIS 394
Query 132 QGVGTNYANWYTN-GTCEMYDMVPTCFTLAPNQFSFTSLGSADPSTAELPPCTEASEGW 189
+GVG N+A + + G C + + P C L + T++GS A PC + G+
Sbjct 395 RGVGMNWATYDKDSGMCALINETPNCLILNAGSIALTAIGSPLEYDAVNYPCHIDTNGY 453
> tpv:TP01_0650 apical membrane antigen 1; K13845 apical merozoite
antigen 1
Length=785
Score = 53.1 bits (126), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 52/191 (27%), Positives = 82/191 (42%), Gaps = 14/191 (7%)
Query 6 KYCSVGDASP-VLTWYCFYPEKTTRP--VSYNSPYVREDHATACPEKAILGAHFGTWDGT 62
KYC A + C YP+K+ + Y + + D CP I + FG++D
Sbjct 405 KYCDKDSADEGTSSLACMYPDKSKDDSHLFYGTSGLHMDWPVVCPVYPIRDSIFGSYDDE 464
Query 63 T--CQRMKAAKQITVPNPTECGKAVFKVSSSD------NPTQYTKPPTTEASSSTSSSNA 114
C + + EC K +F+ S SD N EA ++ S A
Sbjct 465 KDECVPIDPIFVEDADDYEECAKIIFEYSPSDVDISTNNQKLSDVDLYKEAMNNGKLSTA 524
Query 115 VATMWPVGAFSKDEP-RTQGVGTNYANW-YTNGTCEMYDMVPTCFTLAPNQFSFTSLGSA 172
++ M+ +S+D P T+GVG N+A + C + D+VPTC ++ ++ TSL S
Sbjct 525 LSIMF-APRYSEDRPIYTKGVGINWATYSVEEKKCNILDVVPTCLIISNGYYALTSLSSP 583
Query 173 DPSTAELPPCT 183
+ A PC
Sbjct 584 NEDDAINYPCN 594
> tgo:TGME49_115730 apical membrane antigen, putative
Length=388
Score = 51.6 bits (122), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 37/104 (35%), Positives = 48/104 (46%), Gaps = 12/104 (11%)
Query 6 KYCSVGDASPVLTWYCFYPEK---TTRPVSYNSPYVREDH----ATACPEKAILGAHFGT 58
KYCSV LTW CF P K + R + Y S +V E + +ACP A+ A FG
Sbjct 262 KYCSVNGTPSGLTWACFEPVKEKSSARALVYGSAFVAEGNPDAWQSACPNDAVKDALFGK 321
Query 59 WDGTTC---QRMKAAKQITVPNPTECGKAVFK--VSSSDNPTQY 97
W+ C + + N EC K VF + +SD PT Y
Sbjct 322 WEDGQCVPFDTKTSVQSDQATNKEECWKRVFANPLVASDAPTTY 365
> tgo:TGME49_094330 EGF-like domain-containing protein (EC:2.7.10.1
3.1.4.45 3.4.24.18)
Length=1125
Score = 42.4 bits (98), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 45/166 (27%), Positives = 69/166 (41%), Gaps = 24/166 (14%)
Query 19 WYCFYPEKT--TRPVSYNSPYVREDHATACPEKAIL-GAHFGTWD-GTTCQRMKAAKQIT 74
+YC P KT + + Y + + R DH T CP + L +G G C+ M A ++
Sbjct 340 FYCVRPMKTAASSNMVYVTSHTRPDHETKCPPREPLKNVRWGVVSKGKYCKPMNARASLS 399
Query 75 VPNPTECGKAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATMWPVGAFSKDEPRTQGV 134
+CG+ +F +SS+D + ++ ++ VAT +G R
Sbjct 400 NATAEQCGQRLFMLSSADGSSLSSQVRGYHWAT------FVATDCNMGESCAATAR---- 449
Query 135 GTNYANWYTNGTCEMYDMVPTCFTLAPNQFSFTSLGSADPSTAELP 180
G C Y VP C +P +FTSL + DPS A P
Sbjct 450 ----------GKCFFYSTVPECLIHSPTTMAFTSLSAVDPSIAIDP 485
> pfa:PF11_0486 MAEBL, putative
Length=2055
Score = 39.7 bits (91), Expect = 0.012, Method: Composition-based stats.
Identities = 42/155 (27%), Positives = 62/155 (40%), Gaps = 30/155 (19%)
Query 21 CFYPEKTT--RPVSYNSPYVREDHATACPEKAILGAH-FGTWDGTT--CQR-MKAAKQIT 74
C KTT + +Y S ++R D+ T CP + L + FGT+D T C+ M A ++
Sbjct 867 CQILRKTTDSKDWTYVSSFIRPDYETKCPPRYPLKSKVFGTFDQKTGKCKSLMDKAYEVG 926
Query 75 VPNPTECGKAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATMWPVGAFSKDEPRTQGV 134
+ + C + +F VS D S N +W S +E +
Sbjct 927 INKFSVCLEYLFLVSPKD--------------LYNSGRNNYWGIW-AADHSVNENNIE-- 969
Query 135 GTNYANWYTNGTCEMYDMVPTCFTLAPNQFSFTSL 169
NG C + PTC N FSFT+L
Sbjct 970 -------IANGKCYHLVVKPTCVIDKENHFSFTAL 997
Score = 38.5 bits (88), Expect = 0.021, Method: Composition-based stats.
Identities = 34/154 (22%), Positives = 62/154 (40%), Gaps = 26/154 (16%)
Query 21 CFYPEKTTR--PVSYNSPYVREDHATACPEKAILG-AHFGTWDGTT--CQRMKAAKQITV 75
CF P K + +Y S ++R D+ T CP + L FG ++ +T C+ +
Sbjct 294 CFLPVKKEKGNQWTYASSFIRTDYMTKCPPRFPLNHTMFGYFNYSTGKCETYYMNHEKRT 353
Query 76 PNPTECGKAVFKVSSSDNPTQYTKPPTTEASSSTSSSNAVATMWPVGAFSKDEPRTQGVG 135
+ ++C + +F + ++S+ + +W + + ++
Sbjct 354 LSFSKCIETLFN--------------NIKKQDDVNNSSFLWGVWTIENNANEK------- 392
Query 136 TNYANWYTNGTCEMYDMVPTCFTLAPNQFSFTSL 169
TN A+ G+C PTC N FSFT L
Sbjct 393 TNLASMDNTGSCYFLKKKPTCVLKKENHFSFTIL 426
> hsa:3920 LAMP2, CD107b, LAMPB, LGP110; lysosomal-associated
membrane protein 2; K06528 lysosomal-associated membrane protein
1/2
Length=411
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 51/108 (47%), Gaps = 12/108 (11%)
Query 149 MYDMVPTCFTLAPNQFSF--TSLGSADPSTAELPPCTEASEGWEIYGYCECGDGHSTPWK 206
MY + + F++A N S+ LGS+ E S ++I + D P+
Sbjct 303 MYLVNGSVFSIANNNLSYWDAPLGSSYMCNKE--QTVSVSGAFQINTF----DLRVQPFN 356
Query 207 CENGQWIGG---SDDCNCSSILPVALGVSFGLLVPIAALIAYFIYKKK 251
G++ S D + + ++PVA+GV+ G L+ I I+Y I ++K
Sbjct 357 VTQGKYSTAEECSADSDLNFLIPVAVGVALGFLI-IVVFISYMIGRRK 403
> sce:YDR375C BCS1; Bcs1p; K08900 mitochondrial chaperone BCS1
Length=456
Score = 35.4 bits (80), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query 17 LTWYCFYPEKTTRPVSYNSPYVREDHATACPEKAIL---GAHFGTWDG--TTCQRMKAAK 71
LTW +P++ +R +S + Y++ D+ + + +++ G H+ + G +R ++AK
Sbjct 100 LTWMAKHPQRVSRHLSVRTNYIQHDNGSVSTKFSLVPGPGNHWIRYKGAFILIKRERSAK 159
Query 72 QITVPN 77
I + N
Sbjct 160 MIDIAN 165
> cel:C09B9.7 hypothetical protein
Length=913
Score = 35.0 bits (79), Expect = 0.25, Method: Composition-based stats.
Identities = 18/40 (45%), Positives = 25/40 (62%), Gaps = 1/40 (2%)
Query 213 IGGSDDCNCSSILPVALGVSFGLLVPIAALIAYFIYKKKK 252
I GS+DC + IL + L + LL+ + A IAY Y+KKK
Sbjct 748 ISGSEDCGIT-ILHIILSIVTTLLLALIAFIAYVFYQKKK 786
> mmu:110893 Slc8a3, AW742262, MGC90626, Ncx3; solute carrier
family 8 (sodium/calcium exchanger), member 3; K05849 solute
carrier family 8 (sodium/calcium exchanger)
Length=921
Score = 32.0 bits (71), Expect = 2.0, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 24/52 (46%), Gaps = 0/52 (0%)
Query 196 ECGDGHSTPWKCENGQWIGGSDDCNCSSILPVALGVSFGLLVPIAALIAYFI 247
E GD P +N + GS DC ILP+ + L IA +I YF+
Sbjct 31 EAGDSGDVPSAGQNNESCSGSSDCKEGVILPIWYPENPSLGDKIARVIVYFV 82
> tpv:TP04_0666 hypothetical protein; K12862 pleiotropic regulator
1
Length=521
Score = 30.8 bits (68), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 4/37 (10%)
Query 192 YGYCECGDGHSTPWKCENGQWI----GGSDDCNCSSI 224
Y +C C + WKC GQ+I G + NCS+I
Sbjct 390 YSFCSCASDNVKVWKCPEGQFIRNITGHNSILNCSAI 426
> tgo:TGME49_007480 hypothetical protein
Length=5047
Score = 30.4 bits (67), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 29/69 (42%), Gaps = 6/69 (8%)
Query 190 EIYGYCECGDGHSTPWK------CENGQWIGGSDDCNCSSILPVALGVSFGLLVPIAALI 243
++ + EC GH+ P C G + S CS + P + G G L P+A
Sbjct 476 DMKTFHECSPGHACPAAAIFETPCRAGSYQLASGGAACSLVAPGSAGTGLGALRPVACPP 535
Query 244 AYFIYKKKK 252
+F ++ K
Sbjct 536 GHFCVQEAK 544
Lambda K H
0.315 0.131 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9084709576
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40