bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2656_orf2
Length=273
Score E
Sequences producing significant alignments: (Bits) Value
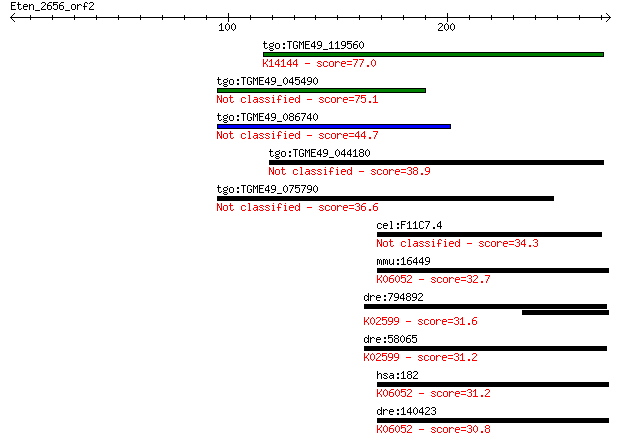
tgo:TGME49_119560 microneme protein MIC3 (EC:3.4.21.6); K14144... 77.0 7e-14
tgo:TGME49_045490 microneme protein 8 (EC:2.7.10.1 3.4.21.22 3... 75.1 2e-13
tgo:TGME49_086740 microneme protein, putative (EC:3.4.21.6) 44.7 3e-04
tgo:TGME49_044180 microneme protein, putative (EC:3.4.21.22 3.... 38.9 0.019
tgo:TGME49_075790 microneme protein, putative (EC:3.4.21.6) 36.6 0.096
cel:F11C7.4 crb-1; Drosophila CRumBs homolog family member (cr... 34.3 0.53
mmu:16449 Jag1, Htu, Ozz, Ser-1; jagged 1; K06052 jagged 32.7 1.4
dre:794892 notch1b, etID309871.5, wu:fc55e11; notch homolog 1b... 31.6 3.1
dre:58065 notch2, notch6, sb:cb884; notch homolog 2; K02599 Notch 31.2 3.8
hsa:182 JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1, MGC104644; jag... 31.2 4.1
dre:140423 jag1b, jag3; jagged 1b; K06052 jagged 30.8 5.9
> tgo:TGME49_119560 microneme protein MIC3 (EC:3.4.21.6); K14144
microneme protein
Length=359
Score = 77.0 bits (188), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 56/182 (30%), Positives = 77/182 (42%), Gaps = 27/182 (14%)
Query 116 GLHCPPDSCCSKTVIEQGKIGNWCSQSWVFCSTAIIYDPAYSY-GTCKCESYRNRCPIGS 174
G CP +CCSKT+ G G +CS +W+FCS+++IY P SY G C CE +RC +
Sbjct 99 GASCPKSACCSKTMCGPGGCGEFCSSNWIFCSSSLIYHPDKSYGGDCSCEKQGHRCDKNA 158
Query 175 SC---LDSSFGAFCKCNEDL-------------EERNGGCHFTTTTTTTTTTTTTTTTTT 218
C LD+ G CKC + + N C T + + T T
Sbjct 159 ECVENLDAGGGVHCKCKDGFVGTGLTCSEDPCSKRGNAKCGPNGTCIVVDSVSYTCTCGD 218
Query 219 TTTTTTTPPARSLCK-------VDDCRPGFC---DVIDNKVVCTCVGGYVSKKLSEEKEQ 268
T P CK ++C PG C +N C C GY + S+ +E
Sbjct 219 GETLVNLPEGGQGCKRTGCHAFRENCSPGRCIDDASHENGYTCECPTGYSREVTSKAEES 278
Query 269 CV 270
CV
Sbjct 279 CV 280
> tgo:TGME49_045490 microneme protein 8 (EC:2.7.10.1 3.4.21.22
3.4.21.6 3.4.21.21)
Length=957
Score = 75.1 bits (183), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 55/98 (56%), Gaps = 3/98 (3%)
Query 95 CFQSLNREACGARASAAEYPGGLHCPPDSCCSKTVIEQGKIGNWCSQSWVFCSTAIIYDP 154
C S +ACG+R + + CP D CCSKT G G+WC +W CS++IIY
Sbjct 334 CRISKKYDACGSREYSDKGLKWTFCPEDFCCSKTACFYGSCGSWCHDNWALCSSSIIYHD 393
Query 155 AYSYGTCKCESYRNRCPIGSSCLDSSF---GAFCKCNE 189
YSYG C C+ ++ C + + C+ ++ GA+C+C E
Sbjct 394 EYSYGKCNCKRFQENCDVNAICVHANREDGGAYCQCKE 431
> tgo:TGME49_086740 microneme protein, putative (EC:3.4.21.6)
Length=721
Score = 44.7 bits (104), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 47/112 (41%), Gaps = 10/112 (8%)
Query 95 CFQSLNREACGARASAAEYPGGLHCPPDSCCSKTVIEQGKIG---NWCSQSWVFCSTAII 151
C S N C +S E CPP CCS + + G G WC + W I
Sbjct 93 CRLSTNNGPCSKNSSDPEIGQWRFCPPGRCCSMS--KCGPAGCSIGWC-EGWGN-PLCWI 148
Query 152 YDPAYSYGTCKCESYRNRCPIGSSCLDSSF---GAFCKCNEDLEERNGGCHF 200
D YS G C CE YR+RC + CL+ + GA+C C + C F
Sbjct 149 KDDTYSDGKCTCEKYRDRCSPNADCLEYNVERGGAYCVCKDGYYGDGKVCEF 200
> tgo:TGME49_044180 microneme protein, putative (EC:3.4.21.22
3.4.21.6)
Length=752
Score = 38.9 bits (89), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 44/156 (28%), Positives = 57/156 (36%), Gaps = 36/156 (23%)
Query 119 CPPDSCCSKTVIEQGKIG-NWCSQSWVFCSTAIIYDPAYSYGTCKCESYRNRCPIGSSC- 176
CP SCCSK G G +WC+ +W + ++ Y C CE Y +C SC
Sbjct 141 CPSGSCCSKVKCAYGSCGGSWCA-AWNS-GLCVFHNGDYESDRCTCEKYGGQCSKNGSCK 198
Query 177 -LDS-SFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTTTTTTPPARSLCKV 234
LD+ GAFC CN T + P C
Sbjct 199 RLDTPEGGAFCYCNPGY------------------------VGDGRTCESDP-----CYG 229
Query 235 DDCRPGFCDVIDNKVVCTCVGGYVSKKLSEEKEQCV 270
C PG C + N VC C GY ++ S +CV
Sbjct 230 SPCAPGTCSRMGNSYVCGCPPGY-TQDTSRGHARCV 264
> tgo:TGME49_075790 microneme protein, putative (EC:3.4.21.6)
Length=614
Score = 36.6 bits (83), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 41/157 (26%), Positives = 59/157 (37%), Gaps = 21/157 (13%)
Query 95 CFQSLNREACGARASAAEYPGGLHCPPDSCCSKTVIEQGKIGNWCSQSWVFCSTA-IIYD 153
C + + E CG + G HCP D CCS K + C++ + C A Y
Sbjct 181 CRNAESGEGCGPYFNVKLNGVGAHCPADECCS-------KYRHGCTKLPLHCEIANTQYL 233
Query 154 PAYSYGTCKCESYRNRCPIGSSCL---DSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTT 210
P +SYG C + +C + C + G +C C + G C T +
Sbjct 234 PDFSYGKCSEAAQCWQCGPNAKCKVVSGRNGGVYCDCLPPFVGKGGNCTLETCQSVNPCG 293
Query 211 TTTTTTTTTTTTTTTPPARSLCKVDDCRPGFCDVIDN 247
T + T T CK C PGF ++DN
Sbjct 294 AGTCVSLENAKTDYT------CK---CNPGF-TLLDN 320
> cel:F11C7.4 crb-1; Drosophila CRumBs homolog family member (crb-1)
Length=1722
Score = 34.3 bits (77), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 44/103 (42%), Gaps = 5/103 (4%)
Query 168 NRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTTTTTTPP 227
N C G +C D+ FC C GG H T +T T T + TT+
Sbjct 410 NPCQNGGTCQDADGQYFCHCTSGF----GGVHCETVDEPSTPIPTLGTFPSFTTSGIDGF 465
Query 228 ARSLCKVDDC-RPGFCDVIDNKVVCTCVGGYVSKKLSEEKEQC 269
++S +DC C +++ VC C GY +K ++ ++C
Sbjct 466 SQSKISCEDCVNSSNCLDVESGPVCICDDGYFGQKCDQKHDKC 508
> mmu:16449 Jag1, Htu, Ozz, Ser-1; jagged 1; K06052 jagged
Length=1218
Score = 32.7 bits (73), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 38/107 (35%), Gaps = 4/107 (3%)
Query 168 NRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTTTTTTPP 227
N C G+ C + + FCKC ED E +N C TTT + T + TP
Sbjct 532 NPCQNGAQCYNRASDYFCKCPEDYEGKN--CSHLKDHCRTTTCEVIDSCTVAMASNDTPE 589
Query 228 ARSLCKVDDCRP-GFCDVID-NKVVCTCVGGYVSKKLSEEKEQCVFN 272
+ C P G C K C C G+ E C N
Sbjct 590 GVRYISSNVCGPHGKCKSQSGGKFTCDCNKGFTGTYCHENINDCESN 636
> dre:794892 notch1b, etID309871.5, wu:fc55e11; notch homolog
1b; K02599 Notch
Length=2465
Score = 31.6 bits (70), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 29/121 (23%), Positives = 47/121 (38%), Gaps = 11/121 (9%)
Query 162 KCESYRNRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTT 221
+C+ N C G +CL++ G C C + + T T +
Sbjct 318 ECDLMPNSCQNGGTCLNTQGGYNCVCVNGWTGDDCSENIDDCADAACHTGATCHDRVASF 377
Query 222 TTTTPPARS--LCKVDD------CRPGF-CDV--IDNKVVCTCVGGYVSKKLSEEKEQCV 270
P R+ LC +DD C+ G CD ++ K +CTC GYV ++ ++C
Sbjct 378 LCECPHGRTGLLCHLDDACISNPCQKGSNCDTNPVNGKAICTCPLGYVGPACDQDVDECS 437
Query 271 F 271
Sbjct 438 L 438
Score = 30.0 bits (66), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 6/45 (13%)
Query 234 VDDCRP------GFCDVIDNKVVCTCVGGYVSKKLSEEKEQCVFN 272
+DDC+P GFC N CTC+ G+ + E+ +C N
Sbjct 888 IDDCKPNPCSNGGFCKDAVNAFTCTCLPGFRGGRCEEDINECESN 932
> dre:58065 notch2, notch6, sb:cb884; notch homolog 2; K02599
Notch
Length=2471
Score = 31.2 bits (69), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 28/121 (23%), Positives = 47/121 (38%), Gaps = 11/121 (9%)
Query 162 KCESYRNRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTT 221
+C N C G +C ++ G C C + + T +T +
Sbjct 299 ECRLQPNACQNGGTCSNTRNGYNCVCVNGWSGLDCSENIDDCAAEPCTAGSTCIDRVASF 358
Query 222 TTTTPPARS--LCKVDD------CRPG-FCDV--IDNKVVCTCVGGYVSKKLSEEKEQCV 270
+ PP ++ LC +DD C+ G CD ++ K C C GY +E+ ++CV
Sbjct 359 VCSCPPGKTGLLCHIDDACISNPCKMGAHCDTNPVNGKFSCNCPSGYKGSTCAEDIDECV 418
Query 271 F 271
Sbjct 419 I 419
> hsa:182 JAG1, AGS, AHD, AWS, CD339, HJ1, JAGL1, MGC104644; jagged
1; K06052 jagged
Length=1218
Score = 31.2 bits (69), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 37/107 (34%), Gaps = 4/107 (3%)
Query 168 NRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTTTTTTPP 227
N C G+ C + + FCKC ED E +N C TT + T + TP
Sbjct 532 NPCQNGAQCYNRASDYFCKCPEDYEGKN--CSHLKDHCRTTPCEVIDSCTVAMASNDTPE 589
Query 228 ARSLCKVDDCRP-GFCDVID-NKVVCTCVGGYVSKKLSEEKEQCVFN 272
+ C P G C K C C G+ E C N
Sbjct 590 GVRYISSNVCGPHGKCKSQSGGKFTCDCNKGFTGTYCHENINDCESN 636
> dre:140423 jag1b, jag3; jagged 1b; K06052 jagged
Length=1213
Score = 30.8 bits (68), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 27/107 (25%), Positives = 39/107 (36%), Gaps = 4/107 (3%)
Query 168 NRCPIGSSCLDSSFGAFCKCNEDLEERNGGCHFTTTTTTTTTTTTTTTTTTTTTTTTTPP 227
N C G+ C + + FCKC +D E +N C TT+ + T + +TP
Sbjct 529 NPCQNGAQCFNLASDYFCKCPDDYEGKN--CSHLKDHCRTTSCQVIDSCTVAVASNSTPE 586
Query 228 ARSLCKVDDCRP-GFC-DVIDNKVVCTCVGGYVSKKLSEEKEQCVFN 272
+ C P G C + C C G+ E C N
Sbjct 587 GVRYISSNVCGPHGRCRSQAGGQFTCECQEGFRGTYCHENINDCESN 633
Lambda K H
0.317 0.130 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10337597784
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40