bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2654_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
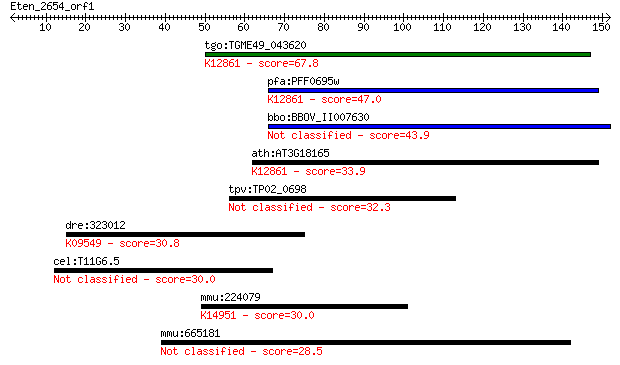
tgo:TGME49_043620 hypothetical protein ; K12861 pre-mRNA-splic... 67.8 1e-11
pfa:PFF0695w conserved Plasmodium protein, unknown function; K... 47.0 3e-05
bbo:BBOV_II007630 18.m06634; hypothetical protein 43.9 2e-04
ath:AT3G18165 MOS4; MOS4 (Modifier of snc1,4); K12861 pre-mRNA... 33.9 0.21
tpv:TP02_0698 hypothetical protein 32.3 0.65
dre:323012 pfdn2, im:6895552, wu:fb79a01, zgc:136473; prefoldi... 30.8 1.6
cel:T11G6.5 hypothetical protein 30.0 2.8
mmu:224079 Atp13a4, 4631413J11Rik, 4832416L12, 9330174J19Rik; ... 30.0 3.3
mmu:665181 Gm11964, OTTMUSG00000005089; predicted gene 11964 28.5
> tgo:TGME49_043620 hypothetical protein ; K12861 pre-mRNA-splicing
factor SPF27
Length=358
Score = 67.8 bits (164), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 44/115 (38%), Positives = 68/115 (59%), Gaps = 19/115 (16%)
Query 50 SAPEFYLTERLPHAMALDALPYVDPLPAEQQQEVQQLLQQEMALIAQE-------AGGPD 102
++P+F L + LP A +D LPYVD L EQ +E + L+QQE+ L+ +E AG
Sbjct 32 ASPKF-LAQALPLAHLVDTLPYVDALTPEQNEEAKSLIQQELVLMNRERRARRQNAGRSS 90
Query 103 N----------LPDYLSD-LPLPKTAILDDAETMLGKEMARKARGEPIPELDLSK 146
+ L +YL + LP P+T LD+ +++G+E+ R RGEP+ +LDLS+
Sbjct 91 DEEASSLEDELLKEYLDELLPAPRTPHLDNPNSLVGRELLRLKRGEPMQKLDLSR 145
> pfa:PFF0695w conserved Plasmodium protein, unknown function;
K12861 pre-mRNA-splicing factor SPF27
Length=255
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 66 LDALPYVDPLPAEQQQEVQQLLQQEMALIAQEAGGPDNLPDYLSDLPLPKTAILDDAETM 125
++ALPY+D E +Q ++L+++EM L+ + + + +YL PLPK L + ++
Sbjct 50 VNALPYIDSYDNELEQNAKRLVEEEMNLMHK----NNEIKNYLETFPLPKITYLSNDNSI 105
Query 126 LGKEMARKARGEPIPELDLSKYT 148
+ E+ R + +L+ Y
Sbjct 106 IQNELKRCEENRKMQKLNFDHYN 128
> bbo:BBOV_II007630 18.m06634; hypothetical protein
Length=195
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 47/88 (53%), Gaps = 4/88 (4%)
Query 66 LDALPYVDPLPAEQQQEVQQLLQQEMALIAQEAGGPDNLPDYLSDLPLPKTAILDDAET- 124
+D+L +VD +P E + +++L+ +E I E G + D L+ P A+ D + +
Sbjct 17 VDSLLFVDAVPVELESRIRELVCEEKRRILDECNGHE--SDVLNKYIEPLGAVPDCSSSG 74
Query 125 -MLGKEMARKARGEPIPELDLSKYTSFT 151
M + + ARGE I LDL KY+ F+
Sbjct 75 HMYHEAVDHCARGEHIQALDLEKYSGFS 102
> ath:AT3G18165 MOS4; MOS4 (Modifier of snc1,4); K12861 pre-mRNA-splicing
factor SPF27
Length=253
Score = 33.9 bits (76), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 32/94 (34%), Positives = 49/94 (52%), Gaps = 18/94 (19%)
Query 62 HAMALDALPYVD-----PLPAEQQQEVQQLLQQEMALIAQEAGGPDNLPDYLSDL-PLPK 115
+A +DALPY+D PL + EV +L+++EM +++ D+L DL PLPK
Sbjct 26 NAEVIDALPYIDDDYGNPLI---KSEVDRLVEEEMRRSSKKPA------DFLKDLPPLPK 76
Query 116 TAILDDAETMLGKEMARKARGEPIPELDL-SKYT 148
+ +LGKE R G+P +D S+Y
Sbjct 77 FDFKNCP--VLGKEYERVRAGKPPVRIDFESRYK 108
> tpv:TP02_0698 hypothetical protein
Length=186
Score = 32.3 bits (72), Expect = 0.65, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 2/59 (3%)
Query 56 LTERLPHAMALDALPYVDPLPAEQQQEVQQLLQQEMALIAQEAGGPDN--LPDYLSDLP 112
L +R + +D+LP+VD +P + +V++L+ EM I E + L +YLSD
Sbjct 10 LYKRNKNYELVDSLPFVDTVPVDLDPKVKELIADEMKSILDENNCQEAELLANYLSDFS 68
> dre:323012 pfdn2, im:6895552, wu:fb79a01, zgc:136473; prefoldin
subunit 2; K09549 prefoldin subunit 2
Length=156
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 29/66 (43%), Gaps = 7/66 (10%)
Query 15 MPASSDLSNGGHS------PGGSQALVHVNGASQEPSNSASSAPEFYLTERLPHAMALDA 68
M A+S S G S P Q + QE + AS A EF + E H++ +D
Sbjct 1 MAANSSNSTSGKSGAKQSTPSAEQVVATFQRMRQEQRSMASKAAEFEM-EINEHSLVIDT 59
Query 69 LPYVDP 74
L VDP
Sbjct 60 LKEVDP 65
> cel:T11G6.5 hypothetical protein
Length=1406
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 14/55 (25%), Positives = 26/55 (47%), Gaps = 0/55 (0%)
Query 12 VPLMPASSDLSNGGHSPGGSQALVHVNGASQEPSNSASSAPEFYLTERLPHAMAL 66
V L +D+ N G QA+ N + +S +P+ Y +R+PH+ ++
Sbjct 564 VQLQLGKTDIENQGTDDVFKQAISRTNAQRESVERESSQSPQLYDPKRIPHSASI 618
> mmu:224079 Atp13a4, 4631413J11Rik, 4832416L12, 9330174J19Rik;
ATPase type 13A4 (EC:3.6.3.-); K14951 cation-transporting
ATPase 13A3/4/5 [EC:3.6.3.-]
Length=1193
Score = 30.0 bits (66), Expect = 3.3, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query 49 SSAPEFYLTERLPHAMALDALPYVDPLPAEQQQEVQQL----LQQEMALIAQEAGG 100
+S +F++ E L H + + V +PAE V Q Q M +I QE GG
Sbjct 563 ASGDDFHIKEMLAHTIVVKPTDMVAQVPAEGLAIVHQFPFSSALQRMTVIVQEMGG 618
> mmu:665181 Gm11964, OTTMUSG00000005089; predicted gene 11964
Length=147
Score = 28.5 bits (62), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 51/104 (49%), Gaps = 8/104 (7%)
Query 39 GASQEPSNSASSAPEFYLTERLPH-AMALDALPYVDPLPAEQQQEVQQLLQQEMALIAQE 97
G +++ S + ++A + TE L H +AL+ + ++ ++ + L Q+++A E
Sbjct 46 GQNKQHSITKNTAKLDWETEELHHDRVALEVGKVI-----QRGRQSKGLTQKDLATKINE 100
Query 98 AGGPDNLPDYLSDLPLPKTAILDDAETMLGKEMARKARGEPIPE 141
P + DY S +P +L E +G ++ K G+PI +
Sbjct 101 K--PQVIADYESGRAIPNNQVLGKTERAIGLKLRGKDVGKPIEK 142
Lambda K H
0.310 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3199347004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40