bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
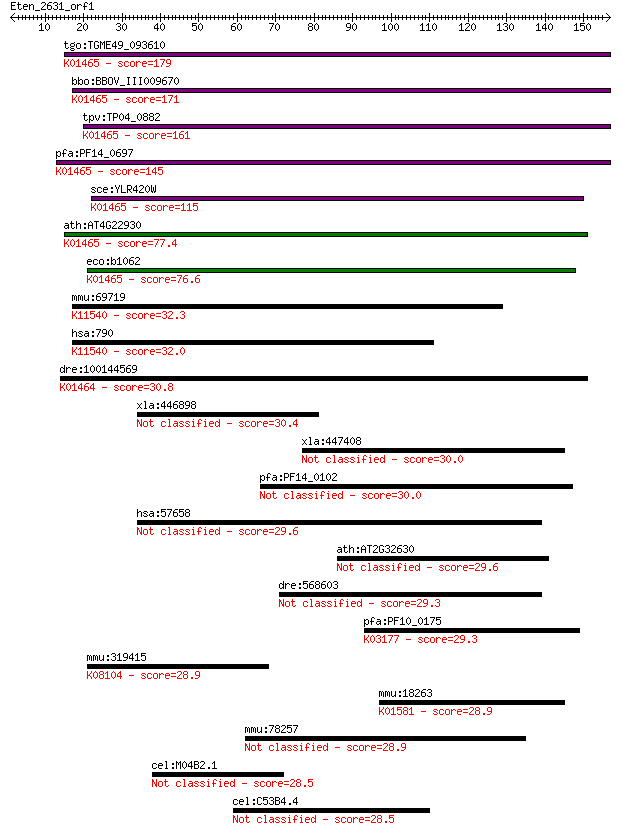
Query= Eten_2631_orf1
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3... 179 3e-45
bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01... 171 7e-43
tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorota... 161 6e-40
pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase ... 145 4e-35
sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymati... 115 8e-26
ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrol... 77.4 2e-14
eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);... 76.6 3e-14
mmu:69719 Cad, 2410008J01Rik, AU018859, Cpad; carbamoyl-phosph... 32.3 0.61
hsa:790 CAD; carbamoyl-phosphate synthetase 2, aspartate trans... 32.0 0.74
dre:100144569 dpys; dihydropyrimidinase (EC:3.5.2.2); K01464 d... 30.8 1.7
xla:446898 mmp3, MGC80992, chds6, mmp-3, mmp13, sl-1, stmy, st... 30.4 2.4
xla:447408 azin1, MGC81852; antizyme inhibitor 1 30.0
pfa:PF14_0102 rhoptry-associated protein 1, RAP1 30.0 3.2
hsa:57658 CALCOCO1, Cocoa, KIAA1536, calphoglin; calcium bindi... 29.6 4.0
ath:AT2G32630 pentatricopeptide (PPR) repeat-containing protein 29.6 4.2
dre:568603 MGC158426, fe06f11, h14c5orf5, wu:fe06f11; zgc:158426 29.3 5.7
pfa:PF10_0175 tRNA pseudouridine synthase, putative; K03177 tR... 29.3 5.9
mmu:319415 Hs3st5, D930005L05Rik, Gm1151, Hs3ost5; heparan sul... 28.9 7.5
mmu:18263 Odc1, MGC103389, ODC; ornithine decarboxylase, struc... 28.9 7.8
mmu:78257 Lrrc9, 4921529O18Rik, 4930432K16Rik; leucine rich re... 28.9 8.0
cel:M04B2.1 mep-1; Mog interacting, Ectopic P granules family ... 28.5 8.2
cel:C53B4.4 hypothetical protein 28.5 9.8
> tgo:TGME49_093610 dihydroorotase protein, putative (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=403
Score = 179 bits (455), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 81/142 (57%), Positives = 105/142 (73%), Gaps = 0/142 (0%)
Query 15 LVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKR 74
LVMP+ DMHTHLRQ E+ + VT I+KGGC VLVMPNT+PP+ TC+QA YR L++
Sbjct 26 LVMPLISDMHTHLRQDEMAEFVTPMIKKGGCRCVLVMPNTIPPVTTCAQAAAYRERLVRI 85
Query 75 DPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKV 134
DPNV+Y+MTL+L E+ ADD+ + AK HV G+K YP+GVTTNS+ GVE QY G+F+
Sbjct 86 DPNVDYMMTLFLSPEVSADDLRQNAKMCHVTGIKSYPKGVTTNSDQGVESYEQYYGIFEA 145
Query 135 MEELGMSLHIHAEKPDAPTLRA 156
M+ELG++LH+H E P L A
Sbjct 146 MQELGLTLHLHGEVPGVAPLDA 167
> bbo:BBOV_III009670 17.m07839; dihydroorotase (EC:3.5.2.3); K01465
dihydroorotase [EC:3.5.2.3]
Length=351
Score = 171 bits (434), Expect = 7e-43, Method: Compositional matrix adjust.
Identities = 81/140 (57%), Positives = 101/140 (72%), Gaps = 0/140 (0%)
Query 17 MPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDP 76
+P+ADD+H HLRQ ELM +VT IR GGC+RVLVMPNT PPI C+QA EYR +L++ +P
Sbjct 3 IPLADDLHCHLRQGELMSLVTPLIRHGGCNRVLVMPNTNPPITNCAQAGEYRRQLMQIEP 62
Query 77 NVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVME 136
V YLMTLYL E+ DDI AK +HV G+K YP G+TTNSE G L +Y +F ME
Sbjct 63 RVTYLMTLYLSPEVSTDDIRLNAKANHVQGIKCYPVGMTTNSEHGFNSLEEYYPLFAEME 122
Query 137 ELGMSLHIHAEKPDAPTLRA 156
LG+SLHIH E+P + LR+
Sbjct 123 RLGLSLHIHGEQPGSNPLRS 142
> tpv:TP04_0882 dihydroorotase (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=295
Score = 161 bits (408), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 80/143 (55%), Positives = 100/143 (69%), Gaps = 6/143 (4%)
Query 20 ADDMHTHLRQVELMDMVTLQIRKGGC------DRVLVMPNTVPPIVTCSQALEYRNELLK 73
ADD+H HLRQ ELM+ V +RKGG +RVLVMPNTVP I +CSQALEYR +LL+
Sbjct 6 ADDLHCHLRQGELMEKVVKYVRKGGWFVLNTYNRVLVMPNTVPEITSCSQALEYRKKLLE 65
Query 74 RDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFK 133
DPNV YLMTLYL ++ ++D+ AK+ HV GVK YP GVTTNS+ G +Y +F
Sbjct 66 LDPNVEYLMTLYLTDDLSSEDLRNNAKKCHVQGVKCYPSGVTTNSDRGFNSFEKYYPLFS 125
Query 134 VMEELGMSLHIHAEKPDAPTLRA 156
ME++G+SLH+H E P AP L A
Sbjct 126 EMEKIGVSLHLHGELPGAPPLTA 148
> pfa:PF14_0697 dihydroorotase, putative; K01465 dihydroorotase
[EC:3.5.2.3]
Length=358
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 69/144 (47%), Positives = 92/144 (63%), Gaps = 0/144 (0%)
Query 13 GRLVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELL 72
+P+ADDMH HLRQ +++D IR+GGC+RVLVMPNT P I TCS A +Y +L
Sbjct 3 NYFYIPIADDMHCHLRQGDMLDFTVNSIRRGGCNRVLVMPNTHPIISTCSDAQKYLYQLK 62
Query 73 KRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVF 132
RD ++ YLMTLYL + D +DI + ++ GVK+YP VTTNS G+ L Y VF
Sbjct 63 SRDDDIEYLMTLYLNKNTDENDILSNYYKCNLQGVKIYPSNVTTNSSDGITSLEPYYKVF 122
Query 133 KVMEELGMSLHIHAEKPDAPTLRA 156
+E+L S+HIH E+P+ L A
Sbjct 123 HALEKLNKSIHIHCEEPNINPLYA 146
> sce:YLR420W URA4; Dihydroorotase, catalyzes the third enzymatic
step in the de novo biosynthesis of pyrimidines, converting
carbamoyl-L-aspartate into dihydroorotate (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=364
Score = 115 bits (287), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 54/130 (41%), Positives = 81/130 (62%), Gaps = 2/130 (1%)
Query 22 DMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNYL 81
DMH H+R+ + ++VT +IR GG +MPN PPI T + +EY+ L K P +L
Sbjct 12 DMHVHVREGAMCELVTPKIRDGGVSIAYIMPNLQPPITTLDRVIEYKKTLQKLAPKTTFL 71
Query 82 MTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVE--DLTQYEGVFKVMEELG 139
M+ YL +++ D I + A++ + GVK YP GVTTNS AGV+ D + + +FK M+E
Sbjct 72 MSFYLSKDLTPDLIHEAAQQHAIRGVKCYPAGVTTNSAAGVDPNDFSAFYPIFKAMQEEN 131
Query 140 MSLHIHAEKP 149
+ L++H EKP
Sbjct 132 LVLNLHGEKP 141
> ath:AT4G22930 PYR4; PYR4 (PYRIMIDIN 4); dihydroorotase/ hydrolase/
hydrolase, acting on carbon-nitrogen (but not peptide)
bonds, in cyclic amides (EC:3.5.2.3); K01465 dihydroorotase
[EC:3.5.2.3]
Length=377
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 11/143 (7%)
Query 15 LVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKR 74
L + DD H HLR +L+ V R +VMPN PP+ + + A+ YR ++K
Sbjct 35 LTITQPDDWHLHLRDGDLLHAVVPH-SASNFKRAIVMPNLKPPVTSTAAAIIYRKFIMKA 93
Query 75 DPN---VNYLMTLYLCREIDADDIAKRAKESHVV-GVKLYPRGVTTNSEAGVEDLTQYEG 130
P+ + LMTLYL + ++I + A+ES VV VKLYP G TTNS+ GV DL +
Sbjct 94 LPSESSFDPLMTLYLTDKTLPEEI-RLARESGVVYAVKLYPAGATTNSQDGVTDL--FGK 150
Query 131 VFKVMEEL---GMSLHIHAEKPD 150
V+EE+ M L +H E D
Sbjct 151 CLPVLEEMVKQNMPLLVHGEVTD 173
> eco:b1062 pyrC, ECK1047, JW1049; dihydro-orotase (EC:3.5.2.3);
K01465 dihydroorotase [EC:3.5.2.3]
Length=348
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 47/133 (35%), Positives = 66/133 (49%), Gaps = 10/133 (7%)
Query 21 DDMHTHLRQVELMDMV---TLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDP- 76
DD H HLR +++ V T +I R +VMPN PP+ T A+ YR +L P
Sbjct 14 DDWHLHLRDGDMLKTVVPYTSEIYG----RAIVMPNLAPPVTTVEAAVAYRQRILDAVPA 69
Query 77 --NVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKV 134
+ LMT YL +D +++ + E KLYP TTNS GV + V +
Sbjct 70 GHDFTPLMTCYLTDSLDPNELERGFNEGVFTAAKLYPANATTNSSHGVTSIDAIMPVLER 129
Query 135 MEELGMSLHIHAE 147
ME++GM L +H E
Sbjct 130 MEKIGMPLLVHGE 142
> mmu:69719 Cad, 2410008J01Rik, AU018859, Cpad; carbamoyl-phosphate
synthetase 2, aspartate transcarbamylase, and dihydroorotase
(EC:2.1.3.2); K11540 carbamoyl-phosphate synthase / aspartate
carbamoyltransferase / dihydroorotase [EC:6.3.5.5 2.1.3.2
3.5.2.3]
Length=2225
Score = 32.3 bits (72), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 33/117 (28%), Positives = 51/117 (43%), Gaps = 11/117 (9%)
Query 17 MPMADDMHTHLRQ-----VELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNEL 71
+P D+H HLR+ E T GG V MPNT PPI+ + AL +L
Sbjct 1464 LPGLIDVHVHLREPGGTHKEDFASGTAAALAGGVTMVCAMPNTRPPIID-APALALAQKL 1522
Query 72 LKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQY 128
+ ++ + L E +A + A + G+KLY T SE ++ + Q+
Sbjct 1523 AEAGARCDFTLFLGASSE-NAGTLGAVAGSA--AGLKLYLN--ETFSELRLDSVAQW 1574
> hsa:790 CAD; carbamoyl-phosphate synthetase 2, aspartate transcarbamylase,
and dihydroorotase (EC:2.1.3.2 3.5.2.3 6.3.5.5);
K11540 carbamoyl-phosphate synthase / aspartate carbamoyltransferase
/ dihydroorotase [EC:6.3.5.5 2.1.3.2 3.5.2.3]
Length=2225
Score = 32.0 bits (71), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 43/99 (43%), Gaps = 9/99 (9%)
Query 17 MPMADDMHTHLRQ-----VELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNEL 71
+P D+H HLR+ E T GG V MPNT PPI+ + AL +L
Sbjct 1464 LPGLIDVHVHLREPGGTHKEDFASGTAAALAGGITMVCAMPNTRPPIID-APALALAQKL 1522
Query 72 LKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLY 110
+ ++ + L E +A + A + G+KLY
Sbjct 1523 AEAGARCDFALFLGASSE-NAGTLGTVAGSA--AGLKLY 1558
> dre:100144569 dpys; dihydropyrimidinase (EC:3.5.2.2); K01464
dihydropyrimidinase [EC:3.5.2.2]
Length=500
Score = 30.8 bits (68), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 36/144 (25%), Positives = 56/144 (38%), Gaps = 10/144 (6%)
Query 14 RLVMPMADDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNT-----VPPIVTCSQALEYR 68
+LV+P D HTH+ ++ M T + G + T V CS Y
Sbjct 53 KLVLPGGIDSHTHM-ELSFMGTTTADDFQTGTQAAVAGGTTMILDFVIAQKGCSLLEAYE 111
Query 69 NELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVV--GVKLYPRGVTTNSEAGVEDLT 126
DP V +L++ DD K+ E+ V G+ + + ++DL
Sbjct 112 RWRRTADPKVCCDYSLHMAVTW-WDDTVKKEMETLVSEKGINSFKMFMAYKDLYMLQDLE 170
Query 127 QYEGVFKVMEELGMSLHIHAEKPD 150
Y VF +E+G +HAE D
Sbjct 171 LY-AVFSTCKEIGAIAQVHAENGD 193
> xla:446898 mmp3, MGC80992, chds6, mmp-3, mmp13, sl-1, stmy,
stmy1, str1; matrix metallopeptidase 3 (stromelysin 1, progelatinase)
(EC:3.4.24.17)
Length=458
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 34 DMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNY 80
D+ +Q G + + PN +PP++T A +N+ K DPN+ +
Sbjct 232 DIAGIQTLYGRRNSIEPKPNPMPPVITPKPAPPPKNQPNKCDPNLQF 278
> xla:447408 azin1, MGC81852; antizyme inhibitor 1
Length=443
Score = 30.0 bits (66), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 21/68 (30%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query 77 NVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVME 136
N+ + TL CR + D AK VVGVK + + N + + L+ VF + +
Sbjct 167 NMTFGTTLKNCRHLL--DCAKELS-VEVVGVKFHVSSSSNNPQTYIHALSDARCVFDMAK 223
Query 137 ELGMSLHI 144
ELG ++I
Sbjct 224 ELGFKMNI 231
> pfa:PF14_0102 rhoptry-associated protein 1, RAP1
Length=782
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 11/88 (12%)
Query 66 EYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGV-------TTNS 118
E+ N+LLK PN++YL TL ++ +DI + SHV L P+ +
Sbjct 424 EFENDLLKFHPNIDYL-TLADGYKLQKNDIYEL---SHVNFCLLNPKTLEEFLKKKEIKD 479
Query 119 EAGVEDLTQYEGVFKVMEELGMSLHIHA 146
G +DL +Y+ F + ++ HI +
Sbjct 480 LMGGDDLIKYKENFDNFMSISITCHIES 507
> hsa:57658 CALCOCO1, Cocoa, KIAA1536, calphoglin; calcium binding
and coiled-coil domain 1
Length=606
Score = 29.6 bits (65), Expect = 4.0, Method: Composition-based stats.
Identities = 27/105 (25%), Positives = 46/105 (43%), Gaps = 18/105 (17%)
Query 34 DMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEYRNELLKRDPNVNYLMTLYLCREIDAD 93
++VTL+ GG D +LV+P +A +N+L + N LM L L E
Sbjct 93 ELVTLEEADGGSDILLVVP----------KATVLQNQLDESQQERNDLMQLKLQLEGQVT 142
Query 94 DIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVMEEL 138
++ R +E R + T + E + QY+G+ + E+
Sbjct 143 ELRSRVQE--------LERALATARQEHTELMEQYKGISRSHGEI 179
> ath:AT2G32630 pentatricopeptide (PPR) repeat-containing protein
Length=624
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 2/57 (3%)
Query 86 LCREIDADDIAKRAKESHVVGVK--LYPRGVTTNSEAGVEDLTQYEGVFKVMEELGM 140
LCR + + K KE V G+K Y N+ D + EGV KVM++ G+
Sbjct 234 LCRRGEVEKSKKLIKEFSVKGIKPEAYTYNTIINAYVKQRDFSGVEGVLKVMKKDGV 290
> dre:568603 MGC158426, fe06f11, h14c5orf5, wu:fe06f11; zgc:158426
Length=678
Score = 29.3 bits (64), Expect = 5.7, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 71 LLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTT-NSEAGVEDLTQYE 129
L++ P VNY + +LCR + +A +ES +G G + + VEDL + E
Sbjct 135 FLQQLPQVNYSLLRFLCRFLSG--VASLQEESWSLGALAAVFGPDIFHLDTDVEDLKEQE 192
Query 130 GVFKVMEEL 138
V +++ EL
Sbjct 193 SVSRILAEL 201
> pfa:PF10_0175 tRNA pseudouridine synthase, putative; K03177
tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=560
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 27/56 (48%), Gaps = 0/56 (0%)
Query 93 DDIAKRAKESHVVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVMEELGMSLHIHAEK 148
D K KE HV+ KL + + TN + + + QY ++ G+ L+ +A K
Sbjct 415 DREGKIIKEEHVIHKKLKKQDILTNLQKFIGHIKQYPPIYSAKRFKGLRLYEYARK 470
> mmu:319415 Hs3st5, D930005L05Rik, Gm1151, Hs3ost5; heparan sulfate
(glucosamine) 3-O-sulfotransferase 5 (EC:2.8.2.23); K08104
[heparan sulfate]-glucosamine 3-sulfotransferase 5 [EC:2.8.2.23]
Length=346
Score = 28.9 bits (63), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 21 DDMHTHLRQVELMDMVTLQIRKGGCDRVLVMPNTVPPIVTCSQALEY 67
+ +H H +L + + +RKGG +L M N P +V SQ + +
Sbjct 79 EQVHLHDLVQQLPKAIIIGVRKGGTRALLEMLNLHPAVVKASQEIHF 125
> mmu:18263 Odc1, MGC103389, ODC; ornithine decarboxylase, structural
1 (EC:4.1.1.17); K01581 ornithine decarboxylase [EC:4.1.1.17]
Length=461
Score = 28.9 bits (63), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 28/50 (56%), Gaps = 2/50 (4%)
Query 97 KRAKESH--VVGVKLYPRGVTTNSEAGVEDLTQYEGVFKVMEELGMSLHI 144
+RAKE + V+GV + T+ E V+ ++ VF + E+G S+H+
Sbjct 182 ERAKELNIDVIGVSFHVGSGCTDPETFVQAVSDARCVFDMATEVGFSMHL 231
> mmu:78257 Lrrc9, 4921529O18Rik, 4930432K16Rik; leucine rich
repeat containing 9
Length=1456
Score = 28.9 bits (63), Expect = 8.0, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 5/73 (6%)
Query 62 SQALEYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKLYPRGVTTNSEAG 121
SQ L++ +E+LK +P + L E +AK SH+V + L+ NS +
Sbjct 644 SQDLKFDDEVLKMEPRIKPRPKLISLDEKTIISLAKTNIYSHIVNLNLH-----GNSLSK 698
Query 122 VEDLTQYEGVFKV 134
+ DL + G+ K+
Sbjct 699 LRDLAKLTGLRKL 711
> cel:M04B2.1 mep-1; Mog interacting, Ectopic P granules family
member (mep-1)
Length=870
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 38 LQIRKGGCDRVLVMPNTV--PPIVTCSQALEYRNEL 71
L +RK D+VLV+PNT+ PP C +E+ E+
Sbjct 287 LSVRKLILDKVLVLPNTISFPPSQVCDLLIEHDPEM 322
> cel:C53B4.4 hypothetical protein
Length=1586
Score = 28.5 bits (62), Expect = 9.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 2/51 (3%)
Query 59 VTCSQALEYRNELLKRDPNVNYLMTLYLCREIDADDIAKRAKESHVVGVKL 109
VT Q +N LL D N NY +Y I++ I + ++ HVV V L
Sbjct 454 VTVLQCSRLKNALLLDDKNKNY--EVYCTVSIESRPIVQNEEQGHVVNVLL 502
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3451799900
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40