bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2572_orf2
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
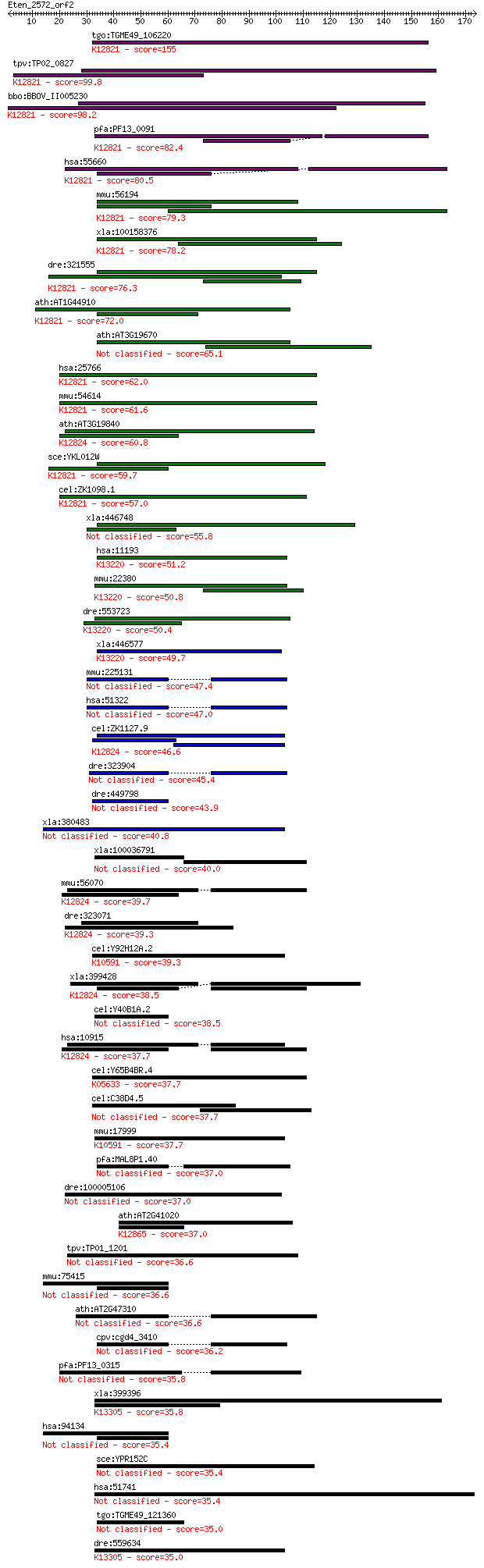
tgo:TGME49_106220 formin binding protein, putative ; K12821 pr... 155 5e-38
tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing... 99.8 4e-21
bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12... 98.2 1e-20
pfa:PF13_0091 conserved Plasmodium protein, unknown function; ... 82.4 6e-16
hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-... 80.5 3e-15
mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40 ... 79.3 7e-15
xla:100158376 prpf40a, fbp-11, fbp11, flaf1, fnbp3, hip10, hyp... 78.2 1e-14
dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe... 76.3 5e-14
ath:AT1G44910 protein binding; K12821 pre-mRNA-processing fact... 72.0 1e-12
ath:AT3G19670 protein binding 65.1 1e-10
hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40 h... 62.0 1e-09
mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mR... 61.6 1e-09
ath:AT3G19840 FF domain-containing protein / WW domain-contain... 60.8 2e-09
sce:YKL012W PRP40; Prp40p; K12821 pre-mRNA-processing factor 40 59.7
cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing ... 57.0 3e-08
xla:446748 wbp4, MGC78936; WW domain binding protein 4 (formin... 55.8 7e-08
hsa:11193 WBP4, FBP21, MGC117310; WW domain binding protein 4 ... 51.2 2e-06
mmu:22380 Wbp4, AW545037, BB101031, FBP21; WW domain binding p... 50.8 2e-06
dre:553723 MGC112384; zgc:112384; K13220 WW domain-binding pro... 50.4 3e-06
xla:446577 MGC81630 protein; K13220 WW domain-binding protein 4 49.7
mmu:225131 Wac, 1110067P07Rik, A230035H12Rik, AI256735, AI2567... 47.4 3e-05
hsa:51322 WAC, BM-016, MGC10753, PRO1741, Wwp4, bA48B24, bA48B... 47.0 3e-05
cel:ZK1127.9 hypothetical protein; K12824 transcription elonga... 46.6 4e-05
dre:323904 waca, wac, wu:fc13f04, wu:fj68f01, zgc:55755; WW do... 45.4 8e-05
dre:449798 wacb, zgc:101828; WW domain containing adaptor with... 43.9 3e-04
xla:380483 arhgap12, MGC52929; Rho GTPase activating protein 12 40.8
xla:100036791 gas7; growth arrest-specific 7 40.0
mmu:56070 Tcerg1, 2410022J09Rik, 2900090C16Rik, AI428505, CA15... 39.7 0.005
dre:323071 tcerg1a, tcerg1, wu:fb80g01; transcription elongati... 39.3 0.006
cel:Y92H12A.2 hypothetical protein; K10591 E3 ubiquitin-protei... 39.3 0.006
xla:399428 tcerg1, MGC130790; potential gravity-related protei... 38.5 0.011
cel:Y40B1A.2 hypothetical protein 38.5 0.012
hsa:10915 TCERG1, CA150, MGC133200, TAF2S, Urn1; transcription... 37.7 0.017
cel:Y65B4BR.4 wwp-1; WW domain Protein (E3 ubiquitin ligase) f... 37.7 0.019
cel:C38D4.5 tag-325; Temporarily Assigned Gene name family mem... 37.7 0.020
mmu:17999 Nedd4, AA959633, AL023035, AU019897, E430025J12Rik, ... 37.7 0.020
pfa:MAL8P1.40 RNA binding protein, putative 37.0 0.029
dre:100005106 plekha7b; pleckstrin homology domain containing,... 37.0 0.029
ath:AT2G41020 WW domain-containing protein; K12865 polyglutami... 37.0 0.034
tpv:TP01_1201 hypothetical protein 36.6 0.039
mmu:75415 Arhgap12, 2810011M08Rik; Rho GTPase activating prote... 36.6 0.040
ath:AT2G47310 flowering time control protein-related / FCA gam... 36.6 0.049
cpv:cgd4_3410 RNA binding protein 36.2 0.061
pfa:PF13_0315 RNA binding protein, putative 35.8 0.065
xla:399396 nedd4l, nedd4, nedd4-2; neural precursor cell expre... 35.8 0.082
hsa:94134 ARHGAP12, DKFZp779N2050, FLJ10971, FLJ20737, FLJ2178... 35.4 0.085
sce:YPR152C URN1; Putative protein of unknown function contain... 35.4 0.092
hsa:51741 WWOX, D16S432E, FOR, FRA16D, HHCMA56, PRO0128, SDR41... 35.4 0.11
tgo:TGME49_121360 RNA recognition motif domain-containing protein 35.0 0.13
dre:559634 nedd4l; neural precursor cell expressed, developmen... 35.0 0.14
> tgo:TGME49_106220 formin binding protein, putative ; K12821
pre-mRNA-processing factor 40
Length=601
Score = 155 bits (393), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 74/124 (59%), Positives = 99/124 (79%), Gaps = 0/124 (0%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHS 91
+GWTEH+ K+GR+YYYN TQ+SQW KP ++ T EE ++ K GW ++ +A+GK YWF S
Sbjct 90 NGWTEHVGKDGRRYYYNAATQQSQWEKPEAMMTEEEKKVYNKLGWIKYSTAEGKEYWFSS 149
Query 92 ATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAKTFLVKLFELKKFPPRISWENASK 151
TK+S W TPKEV+E LK L+EE+E+WP+F +K EA+ ++VKLFELKKFPPRI+WENA K
Sbjct 150 YTKKSTWTTPKEVDEYLKQLEEEKEEWPKFTNKTEARRWIVKLFELKKFPPRINWENAVK 209
Query 152 ILES 155
LE+
Sbjct 210 FLET 213
> tpv:TP02_0827 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=390
Score = 99.8 bits (247), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 52/131 (39%), Positives = 80/131 (61%), Gaps = 6/131 (4%)
Query 28 ITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSY 87
++S WTEH+SK+GRKYYYN T++SQW KP L+T +E I KT W+ F +A+GK +
Sbjct 1 MSSQSLWTEHVSKDGRKYYYNQKTKKSQWEKPNELKTEQELIIEAKTKWRTFATAEGKVF 60
Query 88 WFHSATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAKTFLVKLFELKKFPPRISWE 147
++++ TK SVW P+EV+ +L ED ++ K + E F + +W+
Sbjct 61 YYNTETKESVWEVPEEVKNLLA------EDNLLGTVQDNTKAAFMTFLESFNFTQKTTWD 114
Query 148 NASKILESRPQ 158
NA K+LE+ P+
Sbjct 115 NALKLLEADPK 125
Score = 32.3 bits (72), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 24/70 (34%), Positives = 35/70 (50%), Gaps = 10/70 (14%)
Query 3 QKKKWSRWPVKSEEKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSL 62
QK K S+W +E K + +I + W + EG+ +YYNT T+ES W
Sbjct 22 QKTKKSQWEKPNELKTEQ----ELIIEAKTKWRTFATAEGKVFYYNTETKESVW------ 71
Query 63 QTPEEAQILL 72
+ PEE + LL
Sbjct 72 EVPEEVKNLL 81
> bbo:BBOV_II005230 18.m06432; WW domain containing protein; K12821
pre-mRNA-processing factor 40
Length=457
Score = 98.2 bits (243), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 51/128 (39%), Positives = 83/128 (64%), Gaps = 5/128 (3%)
Query 27 LITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKS 86
+ S WTEH+SK+GR+Y+YN T++SQW KP L+T E +I +T W++F +A+GK
Sbjct 1 MAVSNAYWTEHVSKDGRRYFYNQQTKKSQWEKPDELKTDLERKIESRTNWKQFETAEGKV 60
Query 87 YWFHSATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAKTFLVKLFELKKFPPRISW 146
Y+++S T++SVW+ P+EV +++ + ERE + +KE AK + E F R +W
Sbjct 61 YYYNSVTRQSVWSKPQEVLDVIS--EHERE---ELSTKENAKVAFSRWLEEFNFTRRTTW 115
Query 147 ENASKILE 154
+ A ++LE
Sbjct 116 DMAVRLLE 123
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 55/133 (41%), Gaps = 35/133 (26%)
Query 1 NPQKKKWSRWPVKSEEKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPL 60
N Q KK S+W E K D I S W + + EG+ YYYN++T++S W KP
Sbjct 22 NQQTKK-SQWEKPDELKTD----LERKIESRTNWKQFETAEGKVYYYNSVTRQSVWSKPQ 76
Query 61 ------------SLQTPEEAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEML 108
L T E A++ +EF + T+R+ W+ M
Sbjct 77 EVLDVISEHEREELSTKENAKVAFSRWLEEF-----------NFTRRTTWD-------MA 118
Query 109 KSLKEEREDWPQF 121
L E E WP+F
Sbjct 119 VRLLEVNERWPKF 131
> pfa:PF13_0091 conserved Plasmodium protein, unknown function;
K12821 pre-mRNA-processing factor 40
Length=906
Score = 82.4 bits (202), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 37/85 (43%), Positives = 60/85 (70%), Gaps = 1/85 (1%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSA 92
GW E ++K GRK+YYN++T+ S+W KP L+T EE +I KT W+E+ +DG+ YW+H
Sbjct 260 GWVEMVAKNGRKFYYNSITKCSKWEKPNELKTKEEIRISEKTKWKEYSCSDGRKYWYHEE 319
Query 93 TKRSVWNTPKEVEEM-LKSLKEERE 116
SVW+ P+E++++ L+ E++E
Sbjct 320 KNISVWDEPEEIKKIKLECALEDKE 344
Score = 37.0 bits (84), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 27/38 (71%), Gaps = 0/38 (0%)
Query 118 WPQFKSKEEAKTFLVKLFELKKFPPRISWENASKILES 155
W +F +K +A+ L LFE KK P+++W++A KILE+
Sbjct 431 WEKFDNKNDAREHLKFLFEEKKVNPKMTWDSALKILEA 468
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 73 KTGWQEFCSADGKSYWFHSATKRSVWNTPKEV 104
K GW E + +G+ ++++S TK S W P E+
Sbjct 258 KHGWVEMVAKNGRKFYYNSITKCSKWEKPNEL 289
> hsa:55660 PRPF40A, FBP-11, FBP11, FLAF1, FLJ20585, FNBP3, HIP-10,
HIP10, HYPA, NY-REN-6, Prp40; PRP40 pre-mRNA processing
factor 40 homolog A (S. cerevisiae); K12821 pre-mRNA-processing
factor 40
Length=930
Score = 80.5 bits (197), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 41/86 (47%), Positives = 57/86 (66%), Gaps = 1/86 (1%)
Query 22 IAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCS 81
+AA + + WTEH S +GR YYYNT T++S W KP L+TP E Q+L K W+E+ S
Sbjct 107 VAAGTASGAKSMWTEHKSPDGRTYYYNTETKQSTWEKPDDLKTPAE-QLLSKCPWKEYKS 165
Query 82 ADGKSYWFHSATKRSVWNTPKEVEEM 107
GK Y+++S TK S W PKE+E++
Sbjct 166 DSGKPYYYNSQTKESRWAKPKELEDL 191
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTG 75
W E+ S G+ YYYN+ T+ES+W KP L+ E Q + G
Sbjct 160 WKEYKSDSGKPYYYNSQTKESRWAKPKELEDLEGYQNTIVAG 201
Score = 30.4 bits (67), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 29/56 (51%), Gaps = 5/56 (8%)
Query 112 KEEREDWPQFKS-----KEEAKTFLVKLFELKKFPPRISWENASKILESRPQVVLL 162
KEE E P K+ KEEAK +L + K+ P SWE A K++ + P+ L
Sbjct 349 KEEEESQPAKKTYTWNTKEEAKQAFKELLKEKRVPSNASWEQAMKMIINDPRYSAL 404
> mmu:56194 Prpf40a, 2810012K09Rik, FBP11, Fnbp11, Fnbp3; PRP40
pre-mRNA processing factor 40 homolog A (yeast); K12821 pre-mRNA-processing
factor 40
Length=953
Score = 79.3 bits (194), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 39/74 (52%), Positives = 52/74 (70%), Gaps = 1/74 (1%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
WTEH S +GR YYYNT T++S W KP L+TP E Q+L K W+E+ S GK Y+++S T
Sbjct 146 WTEHKSPDGRTYYYNTETKQSTWEKPDDLKTPAE-QLLSKCPWKEYKSDSGKPYYYNSQT 204
Query 94 KRSVWNTPKEVEEM 107
K S W PKE+E++
Sbjct 205 KESRWAKPKELEDL 218
Score = 38.1 bits (87), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 25/42 (59%), Gaps = 0/42 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTG 75
W E+ S G+ YYYN+ T+ES+W KP L+ E Q + G
Sbjct 187 WKEYKSDSGKPYYYNSQTKESRWAKPKELEDLEGYQNTIVAG 228
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 50/109 (45%), Gaps = 15/109 (13%)
Query 60 LSLQTPEEAQILLKTGWQEFCSADGKSYWFHSATKRSVWN-TPKEVEEMLKSLKEEREDW 118
+++ T E+AQ+ T Q+ + A + +V + TPK KEE E
Sbjct 328 VTVSTEEQAQLANTTAIQDLSGDISSNTGEEPAKQETVSDFTPK---------KEEEESQ 378
Query 119 PQFKS-----KEEAKTFLVKLFELKKFPPRISWENASKILESRPQVVLL 162
P K+ KEEAK +L + K+ P SWE A K++ + P+ L
Sbjct 379 PAKKTYTWNTKEEAKQAFKELLKEKRVPSNASWEQAMKMIINDPRYSAL 427
> xla:100158376 prpf40a, fbp-11, fbp11, flaf1, fnbp3, hip10, hypa,
prp40; PRP40 pre-mRNA processing factor 40 homolog A; K12821
pre-mRNA-processing factor 40
Length=487
Score = 78.2 bits (191), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 40/81 (49%), Positives = 54/81 (66%), Gaps = 1/81 (1%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
WTEH S +GR Y+YN T++S W KP ++TP E Q+L K W+EF S GK Y+++S T
Sbjct 141 WTEHKSPDGRTYFYNAETKQSTWEKPDDMKTPIE-QLLSKCPWKEFKSDSGKPYYYNSQT 199
Query 94 KRSVWNTPKEVEEMLKSLKEE 114
K S W PKE+EE+ +K E
Sbjct 200 KESRWTKPKELEELEVMIKAE 220
Score = 36.2 bits (82), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 35/60 (58%), Gaps = 1/60 (1%)
Query 64 TPEEAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEEREDWPQFKS 123
T E Q K+ W E S DG++Y++++ TK+S W P +++ ++ L + W +FKS
Sbjct 129 TGTEEQTKTKSQWTEHKSPDGRTYFYNAETKQSTWEKPDDMKTPIEQLL-SKCPWKEFKS 187
> dre:321555 prpf40a, fb18h07, fe47a12, fnbp3, wu:fb18h07, wu:fe47a12;
PRP40 pre-mRNA processing factor 40 homolog A (yeast);
K12821 pre-mRNA-processing factor 40
Length=851
Score = 76.3 bits (186), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 39/81 (48%), Positives = 55/81 (67%), Gaps = 1/81 (1%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
WTEH S +G+ YYYNT T++S W KP L++P E Q+L K W+E+ S GK Y+++S T
Sbjct 112 WTEHKSLDGKIYYYNTETKQSTWEKPDELKSPAE-QMLSKCPWKEYKSDTGKPYYYNSQT 170
Query 94 KRSVWNTPKEVEEMLKSLKEE 114
K S W PKE+E++ +K E
Sbjct 171 KESRWTKPKELEDLEAMIKAE 191
Score = 35.8 bits (81), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 39/86 (45%), Gaps = 1/86 (1%)
Query 16 EKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTG 75
EK D + + S W E+ S G+ YYYN+ T+ES+W KP L+ EA I +
Sbjct 135 EKPDELKSPAEQMLSKCPWKEYKSDTGKPYYYNSQTKESRWTKPKELEDL-EAMIKAEEN 193
Query 76 WQEFCSADGKSYWFHSATKRSVWNTP 101
A G + + + SV P
Sbjct 194 GTADVVAPGTTPALTAQNESSVTVAP 219
Score = 34.3 bits (77), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 17/40 (42%), Positives = 25/40 (62%), Gaps = 4/40 (10%)
Query 73 KTGWQEFCSADGKSYWFHSATKRSVWNTPKEV----EEML 108
K+ W E S DGK Y++++ TK+S W P E+ E+ML
Sbjct 109 KSVWTEHKSLDGKIYYYNTETKQSTWEKPDELKSPAEQML 148
> ath:AT1G44910 protein binding; K12821 pre-mRNA-processing factor
40
Length=926
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 56/98 (57%), Gaps = 5/98 (5%)
Query 11 PVKSEEKMDPFIAA--PSLIT--SGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPE 66
PV+ + P + P +T S W EH S +GRKYYYN T++S W KPL L TP
Sbjct 163 PVQQTGQQTPVAVSTDPGNLTPQSASDWQEHTSADGRKYYYNKRTKQSNWEKPLELMTPL 222
Query 67 EAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPKEV 104
E + T W+EF + +GK Y+++ TK S W P+++
Sbjct 223 E-RADASTVWKEFTTPEGKKYYYNKVTKESKWTIPEDL 259
Score = 37.0 bits (84), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 18/38 (47%), Positives = 26/38 (68%), Gaps = 1/38 (2%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTP-EEAQI 70
W E + EG+KYYYN +T+ES+W P L+ E+AQ+
Sbjct 231 WKEFTTPEGKKYYYNKVTKESKWTIPEDLKLAREQAQL 268
> ath:AT3G19670 protein binding
Length=992
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
W EH S +GRKY++N T++S W KP+ L T E + +T W+E S DG+ Y+++ T
Sbjct 209 WVEHTSADGRKYFFNKRTKKSTWEKPVELMTLFE-RADARTDWKEHSSPDGRKYYYNKIT 267
Query 94 KRSVWNTPKEV 104
K+S W P+E+
Sbjct 268 KQSTWTMPEEM 278
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 33/61 (54%), Gaps = 1/61 (1%)
Query 74 TGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAKTFLVK 133
T W E SADG+ Y+F+ TK+S W P E+ + + + R DW + S + K + K
Sbjct 207 TDWVEHTSADGRKYFFNKRTKKSTWEKPVELMTLFERA-DARTDWKEHSSPDGRKYYYNK 265
Query 134 L 134
+
Sbjct 266 I 266
> hsa:25766 PRPF40B, HYPC; PRP40 pre-mRNA processing factor 40
homolog B (S. cerevisiae); K12821 pre-mRNA-processing factor
40
Length=871
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/100 (33%), Positives = 58/100 (58%), Gaps = 6/100 (6%)
Query 20 PFIAAPSLITSGDG-----WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKT 74
P S +G G W+EH++ +GR YYYN ++S W KP L++ E +L +
Sbjct 79 PGADTASSAVAGTGPPRALWSEHVAPDGRIYYYNADDKQSVWEKPSVLKSKAEL-LLSQC 137
Query 75 GWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEE 114
W+E+ S GK Y++++ +K S W PK+++++ +K+E
Sbjct 138 PWKEYKSDTGKPYYYNNQSKESRWTRPKDLDDLEVLVKQE 177
> mmu:54614 Prpf40b, 2610317D23Rik, Hypc, MGC91243; PRP40 pre-mRNA
processing factor 40 homolog B (yeast); K12821 pre-mRNA-processing
factor 40
Length=873
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 32/100 (32%), Positives = 58/100 (58%), Gaps = 6/100 (6%)
Query 20 PFIAAPSLITSGDG-----WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKT 74
P S +G G W+EH++ +GR YYYN ++S W KP L++ E +L +
Sbjct 79 PGADTASSAVAGTGPPRALWSEHVAPDGRIYYYNADDKQSVWEKPSVLKSKAEL-LLSQC 137
Query 75 GWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEE 114
W+E+ S GK Y++++ ++ S W PK+++++ +K+E
Sbjct 138 PWKEYKSDTGKPYYYNNQSQESRWTRPKDLDDLEALVKQE 177
> ath:AT3G19840 FF domain-containing protein / WW domain-containing
protein; K12824 transcription elongation regulator 1
Length=743
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 51/101 (50%), Gaps = 9/101 (8%)
Query 22 IAAPSLITSG-DGWTEHISKEGRKYYYNTLTQESQWGKPLSLQT--------PEEAQILL 72
IA L+ + D WT H S+ G YYYN++T +S + KP P IL
Sbjct 146 IAGSQLVGNRLDAWTAHKSEAGVLYYYNSVTGQSTYEKPPGFGGEPDKVPVQPIPVSILP 205
Query 73 KTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKE 113
T W + DGK Y++++ TK S W P EV++ K L+E
Sbjct 206 GTDWALVSTNDGKKYYYNNKTKVSSWQIPAEVKDFGKKLEE 246
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 20 PFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQ 63
P P I G W + +G+KYYYN T+ S W P ++
Sbjct 195 PVQPIPVSILPGTDWALVSTNDGKKYYYNNKTKVSSWQIPAEVK 238
> sce:YKL012W PRP40; Prp40p; K12821 pre-mRNA-processing factor
40
Length=583
Score = 59.7 bits (143), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 32/84 (38%), Positives = 50/84 (59%), Gaps = 2/84 (2%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
W E GR YYYNTLT++S W KP L + EE +L + GW+ +ADGK Y+++ T
Sbjct 4 WKEAKDASGRIYYYNTLTKKSTWEKPKELISQEEL-LLRENGWKAAKTADGKVYYYNPTT 62
Query 94 KRSVWNTPKEVEEMLKSLKEERED 117
+ + W P E+ ++ + E++ D
Sbjct 63 RETSWTIPA-FEKKVEPIAEQKHD 85
Score = 32.7 bits (73), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 16 EKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
EK I+ L+ +GW + +G+ YYYN T+E+ W P
Sbjct 27 EKPKELISQEELLLRENGWKAAKTADGKVYYYNPTTRETSWTIP 70
> cel:ZK1098.1 hypothetical protein; K12821 pre-mRNA-processing
factor 40
Length=724
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 59/112 (52%), Gaps = 22/112 (19%)
Query 20 PFIAAPS-----LITSGDG-------------WTEHISKEGRKYYYNTLTQESQWGKPLS 61
P +AAP+ L+ G G W+ H +++G YY+N +T+++ W KP
Sbjct 52 PHVAAPTRPSPMLVPPGMGIDESHSSPSVESDWSVHTNEKGTPYYHNRVTKQTSWIKPDV 111
Query 62 LQTPEEAQIL---LKTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKS 110
L+TP E + W+EF S DGK Y++++ TK++ W P + EE+ K
Sbjct 112 LKTPLERSTSGQPQQGQWKEFMSDDGKPYYYNTLTKKTQWVKP-DGEEITKG 162
> xla:446748 wbp4, MGC78936; WW domain binding protein 4 (formin
binding protein 21)
Length=374
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 49/97 (50%), Gaps = 2/97 (2%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTG--WQEFCSADGKSYWFHS 91
W + IS+EG YYYNTLT ESQW +P Q E +G W E S +G +Y++++
Sbjct 121 WKKDISQEGYPYYYNTLTGESQWEEPEGFQDKSEESNKGGSGSVWVEGLSEEGFTYYYNT 180
Query 92 ATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAK 128
T S W P+ L + E+E EEAK
Sbjct 181 KTGESSWEKPENFVSNLPAENAEKEATNTENKSEEAK 217
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 18/33 (54%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 30 SGDGWTEHISKEGRKYYYNTLTQESQWGKPLSL 62
SG W E +S+EG YYYNT T ES W KP +
Sbjct 161 SGSVWVEGLSEEGFTYYYNTKTGESSWEKPENF 193
> hsa:11193 WBP4, FBP21, MGC117310; WW domain binding protein
4 (formin binding protein 21); K13220 WW domain-binding protein
4
Length=376
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSAT 93
W E I+ EG YYY+ ++ SQW KP Q + + +KT W E S DG +Y++++ T
Sbjct 128 WVEGITSEGYHYYYDLISGASQWEKPEGFQG-DLKKTAVKTVWVEGLSEDGFTYYYNTET 186
Query 94 KRSVWNTPKE 103
S W P +
Sbjct 187 GESRWEKPDD 196
> mmu:22380 Wbp4, AW545037, BB101031, FBP21; WW domain binding
protein 4; K13220 WW domain-binding protein 4
Length=376
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 1/71 (1%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHSA 92
GW E ++ +G YYY+ +T SQW KP Q + K W E S DG +Y++++
Sbjct 128 GWVEGVTADGHCYYYDLITGASQWEKPEGFQG-NLKKTAAKAVWVEGLSEDGYTYYYNTE 186
Query 93 TKRSVWNTPKE 103
T S W P++
Sbjct 187 TGESKWEKPED 197
Score = 30.0 bits (66), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 14/37 (37%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 73 KTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLK 109
K GW E +ADG Y++ T S W P+ + LK
Sbjct 126 KGGWVEGVTADGHCYYYDLITGASQWEKPEGFQGNLK 162
> dre:553723 MGC112384; zgc:112384; K13220 WW domain-binding protein
4
Length=412
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 40/79 (50%), Gaps = 7/79 (8%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKP-------LSLQTPEEAQILLKTGWQEFCSADGK 85
W + +G YYYNTLT ESQW KP +S + Q + W E S DG
Sbjct 128 AWVSGTTADGLLYYYNTLTAESQWEKPDGFVDECVSSTAGQTQQESSGSAWMEAVSPDGF 187
Query 86 SYWFHSATKRSVWNTPKEV 104
+Y++++ + S W P+E+
Sbjct 188 TYYYNTESGESSWEKPEEL 206
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 22/36 (61%), Gaps = 0/36 (0%)
Query 29 TSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQT 64
+SG W E +S +G YYYNT + ES W KP L +
Sbjct 173 SSGSAWMEAVSPDGFTYYYNTESGESSWEKPEELSS 208
> xla:446577 MGC81630 protein; K13220 WW domain-binding protein
4
Length=379
Score = 49.7 bits (117), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQ--TPEEAQILLKTGWQEFCSADGKSYWFHS 91
W + IS EG YYYNTLT ES+W +P Q + E + + W E S +G +Y++++
Sbjct 121 WKKDISPEGYPYYYNTLTGESKWEEPEGFQDKSEESNKAGSSSVWVESLSEEGFTYYYNT 180
Query 92 ATKRSVWNTP 101
T S W P
Sbjct 181 KTGESSWEKP 190
> mmu:225131 Wac, 1110067P07Rik, A230035H12Rik, AI256735, AI256776,
Wwp4; WW domain containing adaptor with coiled-coil
Length=543
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/30 (63%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 30 SGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
S D W+EHIS G+KYYYN T+ SQW KP
Sbjct 131 SADDWSEHISSSGKKYYYNCRTEVSQWEKP 160
Score = 28.9 bits (63), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKE 103
W E S+ GK Y+++ T+ S W PKE
Sbjct 135 WSEHISSSGKKYYYNCRTEVSQWEKPKE 162
> hsa:51322 WAC, BM-016, MGC10753, PRO1741, Wwp4, bA48B24, bA48B24.1;
WW domain containing adaptor with coiled-coil
Length=647
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 19/30 (63%), Positives = 22/30 (73%), Gaps = 0/30 (0%)
Query 30 SGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
S D W+EHIS G+KYYYN T+ SQW KP
Sbjct 131 SADDWSEHISSSGKKYYYNCRTEVSQWEKP 160
Score = 28.9 bits (63), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKE 103
W E S+ GK Y+++ T+ S W PKE
Sbjct 135 WSEHISSSGKKYYYNCRTEVSQWEKPKE 162
> cel:ZK1127.9 hypothetical protein; K12824 transcription elongation
regulator 1
Length=905
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 25/94 (26%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLS---LQTPEEAQILLKT---------------- 74
W E + EG+KY+Y+ + + + W +P + + PE AQ++ +
Sbjct 159 WVETETAEGKKYFYHPVNRNTIWERPQNAKIVTQPELAQLIHRATEEEKNREERMPHGQI 218
Query 75 ------GWQEFCSADGKSYWFHSATKRSVWNTPK 102
W EF + DG+ Y+F+S T+ + W PK
Sbjct 219 PQNPDDAWNEFNAPDGRKYYFNSITQENTWEKPK 252
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKPLSL 62
D W E + +GRKYY+N++TQE+ W KP +L
Sbjct 224 DAWNEFNAPDGRKYYFNSITQENTWEKPKAL 254
Score = 29.6 bits (65), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Query 62 LQTPEEAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPK 102
L EE Q L W E +A+GK Y++H + ++W P+
Sbjct 149 LSGCEEGQEL----WVETETAEGKKYFYHPVNRNTIWERPQ 185
> dre:323904 waca, wac, wu:fc13f04, wu:fj68f01, zgc:55755; WW
domain containing adaptor with coiled-coil a
Length=558
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 18/29 (62%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 31 GDGWTEHISKEGRKYYYNTLTQESQWGKP 59
D W+EHIS G+KYYYN T+ SQW KP
Sbjct 123 ADDWSEHISSSGKKYYYNCRTEVSQWEKP 151
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 12/28 (42%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKE 103
W E S+ GK Y+++ T+ S W PKE
Sbjct 126 WSEHISSSGKKYYYNCRTEVSQWEKPKE 153
> dre:449798 wacb, zgc:101828; WW domain containing adaptor with
coiled-coil b
Length=269
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/28 (64%), Positives = 21/28 (75%), Gaps = 0/28 (0%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKP 59
D W+EHIS G+KYYYN T+ SQW KP
Sbjct 120 DDWSEHISSSGKKYYYNCRTEVSQWEKP 147
> xla:380483 arhgap12, MGC52929; Rho GTPase activating protein
12
Length=776
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 40/93 (43%), Gaps = 5/93 (5%)
Query 14 SEEKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTP----EEAQ 69
S+ + P A+P + +G+ W H GR YY+N TQE W P + E+
Sbjct 234 SQSALPPQPASPPVQITGE-WETHKDNSGRYYYFNKTTQERTWKPPRGTKEAGSGKSESS 292
Query 70 ILLKTGWQEFCSADGKSYWFHSATKRSVWNTPK 102
W + G+ Y++ + RS W PK
Sbjct 293 SQADQEWIKHVDDQGRPYYYSADGSRSEWELPK 325
> xla:100036791 gas7; growth arrest-specific 7
Length=474
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 26/37 (70%), Gaps = 4/37 (10%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGK----PLSLQTP 65
GW ++S +GR+YY NTLT E+ W + P+SL++P
Sbjct 81 GWQSYMSPQGRRYYVNTLTNETTWERPNSSPVSLKSP 117
Score = 33.5 bits (75), Expect = 0.41, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 66 EEAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKS 110
++ Q+ L GWQ + S G+ Y+ ++ T + W P LKS
Sbjct 72 DQTQVPLPQGWQSYMSPQGRRYYVNTLTNETTWERPNSSPVSLKS 116
> mmu:56070 Tcerg1, 2410022J09Rik, 2900090C16Rik, AI428505, CA150b,
FBP23, FBP28, Taf2s, ca150, p144; transcription elongation
regulator 1 (CA150); K12824 transcription elongation regulator
1
Length=1100
Score = 39.7 bits (91), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 31/48 (64%), Gaps = 0/48 (0%)
Query 23 AAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI 70
AP+L + + W E+ + +G+ YYYN T+ES W KP ++ +++++
Sbjct 126 GAPALPPTEEIWVENKTPDGKVYYYNARTRESAWTKPDGVKVIQQSEL 173
Score = 37.4 bits (85), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 18/46 (39%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 21 FIAAPSLI---TSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQ 63
A+P+ + T+ WTE+ + +G+ YYYN T ES W KP L+
Sbjct 421 IAASPATLAGATAVSEWTEYKTADGKTYYYNNRTLESTWEKPQELK 466
Score = 29.6 bits (65), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKS 110
W E + DGK Y++++ T+ S W P V+ + +S
Sbjct 137 WVENKTPDGKVYYYNARTRESAWTKPDGVKVIQQS 171
> dre:323071 tcerg1a, tcerg1, wu:fb80g01; transcription elongation
regulator 1a (CA150); K12824 transcription elongation regulator
1
Length=1000
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 28 ITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI 70
+++ + W E+ S EG+ YYYN T+ES W KP ++ ++A +
Sbjct 135 VSTEEIWVENKSPEGKVYYYNARTRESSWTKPEGVKVIQQADL 177
Score = 38.9 bits (89), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 32/65 (49%), Gaps = 3/65 (4%)
Query 22 IAAPSLITSGDG---WTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQE 78
IA P+ + G W E + EG+ YYYN TQE+ W KP L+ E+ + +E
Sbjct 338 IAGPAGLPGALGTSDWAEFKTPEGKSYYYNKHTQETTWDKPEELRDTEKESEKVMDSVEE 397
Query 79 FCSAD 83
A+
Sbjct 398 MMEAE 402
> cel:Y92H12A.2 hypothetical protein; K10591 E3 ubiquitin-protein
ligase NEDD4 [EC:6.3.2.19]
Length=724
Score = 39.3 bits (90), Expect = 0.006, Method: Composition-based stats.
Identities = 20/81 (24%), Positives = 37/81 (45%), Gaps = 11/81 (13%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKP----------LSLQTPEEAQILLKTGWQEFCS 81
DGW ++ GR ++ + T+ + W P L +T +E L GW++
Sbjct 274 DGWDMQVAPNGRTFFIDHRTKTTTWTDPRPGAATRVPLLRGKTDDEIGAL-PAGWEQRVH 332
Query 82 ADGKSYWFHSATKRSVWNTPK 102
ADG+ ++ +R+ W P+
Sbjct 333 ADGRVFFIDHNRRRTQWEDPR 353
> xla:399428 tcerg1, MGC130790; potential gravity-related protein
1; K12824 transcription elongation regulator 1
Length=722
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 36/56 (64%), Gaps = 1/56 (1%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEEREDW-PQFKSKEEAKTF 130
W E+ +ADGK+Y++++ T S W+ P+E++E K +++ +E P+ KEE +
Sbjct 468 WSEYKTADGKTYYYNTRTLESTWDKPQELKEKDKEVEKVKESTKPEAVIKEEPQAM 523
Score = 38.5 bits (88), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 24 APSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI 70
AP+L + + W E+ + +G+ YYYN T+ES W KP ++ +++++
Sbjct 120 APTLPPNEEIWVENKTPDGKVYYYNARTRESAWSKPDGVKVIQQSEL 166
Score = 37.4 bits (85), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQ 63
W+E+ + +G+ YYYNT T ES W KP L+
Sbjct 468 WSEYKTADGKTYYYNTRTLESTWDKPQELK 497
Score = 29.6 bits (65), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKS 110
W E + DGK Y++++ T+ S W+ P V+ + +S
Sbjct 130 WVENKTPDGKVYYYNARTRESAWSKPDGVKVIQQS 164
> cel:Y40B1A.2 hypothetical protein
Length=120
Score = 38.5 bits (88), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 16/27 (59%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKP 59
GWTE +S G+ YYYN T+ SQW KP
Sbjct 10 GWTEQMSSSGKMYYYNKKTEISQWDKP 36
> hsa:10915 TCERG1, CA150, MGC133200, TAF2S, Urn1; transcription
elongation regulator 1; K12824 transcription elongation regulator
1
Length=1077
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 30/48 (62%), Gaps = 0/48 (0%)
Query 23 AAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI 70
P+L + + W E+ + +G+ YYYN T+ES W KP ++ +++++
Sbjct 126 GTPALPPTEEIWVENKTPDGKVYYYNARTRESAWTKPDGVKVIQQSEL 173
Score = 36.6 bits (83), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Query 21 FIAAPSLI---TSGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
A+P+ + T+ WTE+ + +G+ YYYN T ES W KP
Sbjct 398 IAASPATLAGATAVSEWTEYKTADGKTYYYNNRTLESTWEKP 439
Score = 32.3 bits (72), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPK 102
W E+ +ADGK+Y++++ T S W P+
Sbjct 414 WTEYKTADGKTYYYNNRTLESTWEKPQ 440
Score = 29.6 bits (65), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKS 110
W E + DGK Y++++ T+ S W P V+ + +S
Sbjct 137 WVENKTPDGKVYYYNARTRESAWTKPDGVKVIQQS 171
> cel:Y65B4BR.4 wwp-1; WW domain Protein (E3 ubiquitin ligase)
family member (wwp-1); K05633 atrophin-1 interacting protein
5 (WW domain containing E3 ubiquitin protein ligase 1) [EC:6.3.2.19]
Length=794
Score = 37.7 bits (86), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 36/79 (45%), Gaps = 10/79 (12%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSADGKSYWFHS 91
DGW + GRKYY + T+ + W +P + P+ GW+ G+ Y+
Sbjct 223 DGWEMRFDQYGRKYYVDHTTKSTTWERPSTQPLPQ--------GWEMRRDPRGRVYYVDH 274
Query 92 ATKRSVWNTPKEVEEMLKS 110
T+ + W P +ML++
Sbjct 275 NTRTTTWQRP--TADMLEA 291
> cel:C38D4.5 tag-325; Temporarily Assigned Gene name family member
(tag-325)
Length=837
Score = 37.7 bits (86), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 32 DGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILL---KTGWQEFCSADG 84
+GW E+ + GR +++N T +SQW P ++TP + Q L +T CS G
Sbjct 100 NGWFEYETDVGRTFFFNKETGKSQWIPPRFIRTPAQVQEFLRATRTNLDTTCSFQG 155
Score = 29.6 bits (65), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 27/47 (57%), Gaps = 6/47 (12%)
Query 72 LKTGWQEFCSADGKSYWFHSATKRSVW------NTPKEVEEMLKSLK 112
L GW E+ + G++++F+ T +S W TP +V+E L++ +
Sbjct 98 LLNGWFEYETDVGRTFFFNKETGKSQWIPPRFIRTPAQVQEFLRATR 144
> mmu:17999 Nedd4, AA959633, AL023035, AU019897, E430025J12Rik,
Nedd4-1, Nedd4a, mKIAA0093; neural precursor cell expressed,
developmentally down-regulated 4; K10591 E3 ubiquitin-protein
ligase NEDD4 [EC:6.3.2.19]
Length=887
Score = 37.7 bits (86), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 35/83 (42%), Gaps = 13/83 (15%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEE---AQILLKT----------GWQEF 79
GW E GR YY + ++ + W KP P A + KT GW+E
Sbjct 410 GWEEKQDDRGRSYYVDHNSKTTTWSKPTMQDDPRSKIPAHLRGKTDSNDLGPLPPGWEER 469
Query 80 CSADGKSYWFHSATKRSVWNTPK 102
DG+ ++ + K++ W P+
Sbjct 470 THTDGRVFFINHNIKKTQWEDPR 492
> pfa:MAL8P1.40 RNA binding protein, putative
Length=286
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 19/26 (73%), Gaps = 0/26 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKP 59
W +H++ +G YYYN++T SQW KP
Sbjct 103 WQKHVTTDGHPYYYNSVTGHSQWEKP 128
Score = 34.3 bits (77), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 66 EEAQILLKTGWQEFCSADGKSYWFHSATKRSVWNTPKEV 104
+AQ K WQ+ + DG Y+++S T S W PKE+
Sbjct 93 NDAQGGNKNVWQKHVTTDGHPYYYNSVTGHSQWEKPKEM 131
> dre:100005106 plekha7b; pleckstrin homology domain containing,
family A member 7b
Length=1266
Score = 37.0 bits (84), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 42/84 (50%), Gaps = 4/84 (4%)
Query 22 IAAP-SLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQIL---LKTGWQ 77
+AAP T D W+ + ++GR ++ N T+++ W P + + ++ L GW+
Sbjct 1 MAAPLGRDTLPDHWSYGVCRDGRVFFINDKTRDTTWLHPRTGEPVNSGHMIRSDLPRGWE 60
Query 78 EFCSADGKSYWFHSATKRSVWNTP 101
E + +G SY+ + + + + P
Sbjct 61 EGFTEEGASYFINHNQRSTTFRHP 84
> ath:AT2G41020 WW domain-containing protein; K12865 polyglutamine-binding
protein 1
Length=463
Score = 37.0 bits (84), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 32/67 (47%), Gaps = 3/67 (4%)
Query 42 GRKYYYNTLTQESQWGKP--LSLQTPEEAQILLKTGWQE-FCSADGKSYWFHSATKRSVW 98
G YYYN T QW +P LS T +L K W E F A G Y++++ T S W
Sbjct 207 GATYYYNQHTGTCQWERPVELSYATSSAPPVLSKEEWIETFDEASGHKYFYNTRTHVSQW 266
Query 99 NTPKEVE 105
P ++
Sbjct 267 EPPASLQ 273
Score = 34.3 bits (77), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 15/24 (62%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 42 GRKYYYNTLTQESQWGKPLSLQTP 65
G KY+YNT T SQW P SLQ P
Sbjct 252 GHKYFYNTRTHVSQWEPPASLQKP 275
> tpv:TP01_1201 hypothetical protein
Length=268
Score = 36.6 bits (83), Expect = 0.039, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 32/85 (37%), Gaps = 7/85 (8%)
Query 23 AAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQEFCSA 82
+ P++ S W + + EG YYYN T ++QW KP L P G F
Sbjct 97 SVPAVPASSVLWQQFTTAEGVPYYYNVRTGQTQWEKPAELMAPART----VAGGSSFGPP 152
Query 83 DGKSYWFHSATKRSVWNTPKEVEEM 107
+ FH WN VE
Sbjct 153 GANLFVFHVPAN---WNDLDLVEHF 174
> mmu:75415 Arhgap12, 2810011M08Rik; Rho GTPase activating protein
12
Length=838
Score = 36.6 bits (83), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 14 SEEKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
S+ + P +P++ +G+ W H GR YYYN TQE W P
Sbjct 250 SQSALPPLPGSPAIQVNGE-WETHKDSSGRCYYYNRTTQERTWKPP 294
Score = 29.3 bits (64), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKP 59
W +H+ +GR+YYY+ S+W P
Sbjct 361 WLKHVDDQGRQYYYSADGSRSEWELP 386
> ath:AT2G47310 flowering time control protein-related / FCA gamma-related
Length=512
Score = 36.6 bits (83), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 26 SLITSGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
+ ++S W+EH G KYY++ +T ES W KP
Sbjct 399 NTVSSECDWSEHTCPNGNKYYFHCITCESTWEKP 432
Score = 30.0 bits (66), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 18/39 (46%), Gaps = 0/39 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKEE 114
W E +G Y+FH T S W P E + LKE+
Sbjct 407 WSEHTCPNGNKYYFHCITCESTWEKPDEYSMYERWLKEQ 445
> cpv:cgd4_3410 RNA binding protein
Length=906
Score = 36.2 bits (82), Expect = 0.061, Method: Composition-based stats.
Identities = 12/26 (46%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKP 59
W E+ + +G+ YY+N LTQ +QW P
Sbjct 690 WKEYFTSDGKPYYHNELTQVTQWEVP 715
Score = 31.2 bits (69), Expect = 1.6, Method: Composition-based stats.
Identities = 10/28 (35%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKE 103
W+E+ ++DGK Y+ + T+ + W P E
Sbjct 690 WKEYFTSDGKPYYHNELTQVTQWEVPPE 717
> pfa:PF13_0315 RNA binding protein, putative
Length=509
Score = 35.8 bits (81), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 27/58 (46%), Gaps = 13/58 (22%)
Query 20 PFIAAPSLITSGDG-------------WTEHISKEGRKYYYNTLTQESQWGKPLSLQT 64
P I++P+ I G+ W E+ S EGR YYYN T +QW P +T
Sbjct 291 PSISSPNNINFGNNFSVNNNYPRQVGPWKEYYSGEGRPYYYNEQTNTTQWEMPKEFET 348
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 76 WQEFCSADGKSYWFHSATKRSVWNTPKEVEEML 108
W+E+ S +G+ Y+++ T + W PKE E +
Sbjct 318 WKEYYSGEGRPYYYNEQTNTTQWEMPKEFETLF 350
> xla:399396 nedd4l, nedd4, nedd4-2; neural precursor cell expressed,
developmentally down-regulated 4-like; K13305 E3 ubiquitin-protein
ligase NEDD4-like [EC:6.3.2.19]
Length=971
Score = 35.8 bits (81), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 32/144 (22%), Positives = 60/144 (41%), Gaps = 17/144 (11%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI----------LLKTGWQEFCSA 82
GW I+ GR ++ + T+ + W P L+ P + L GW+E
Sbjct 498 GWEMRIAPNGRPFFIDHNTKTTTWEDP-RLKFPVHMRTKASLNPNDLGPLPPGWEERIHM 556
Query 83 DGKSYWFHSATKRSVWNTPKEVEEMLKS-----LKEEREDWPQFKSKEEAKTFLVKLFEL 137
DG++++ TK + W P+ + +E ++ + F+ K + + FE+
Sbjct 557 DGRTFYIDHNTKITQWEDPRLQNPAITGPAVPYSREFKQKYDYFRKKLKKPADIPNRFEM 616
Query 138 KKFPPRISWENASKILE-SRPQVV 160
K I E+ +I+ RP V+
Sbjct 617 KLHRNNIFEESYRRIMSVKRPDVL 640
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 22/46 (47%), Gaps = 0/46 (0%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQILLKTGWQE 78
GW E + GR YY N + +QW +P + E+ ++ QE
Sbjct 195 GWEEKVDNLGRTYYVNHNNKSTQWQRPSLIDVASESDNNIRHIQQE 240
> hsa:94134 ARHGAP12, DKFZp779N2050, FLJ10971, FLJ20737, FLJ21785,
FLJ45709; Rho GTPase activating protein 12
Length=846
Score = 35.4 bits (80), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 14 SEEKMDPFIAAPSLITSGDGWTEHISKEGRKYYYNTLTQESQWGKP 59
S+ + P +P++ +G+ W H GR YYYN TQE W P
Sbjct 252 SQSALPPLPGSPAIQINGE-WETHKDSSGRCYYYNRGTQERTWKPP 296
Score = 29.3 bits (64), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 10/26 (38%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKP 59
W +H+ +GR+YYY+ S+W P
Sbjct 364 WLKHVDDQGRQYYYSADGSRSEWELP 389
> sce:YPR152C URN1; Putative protein of unknown function containing
WW and FF domains; overexpression causes accumulation
of cells in G1 phase
Length=465
Score = 35.4 bits (80), Expect = 0.092, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 45/94 (47%), Gaps = 14/94 (14%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKP-----LSLQT-PEEAQ--------ILLKTGWQEF 79
W E + G+KYYYN T++S+W KP +L++ +E+Q + L GW
Sbjct 5 WQEFKTPAGKKYYYNKNTKQSRWEKPNLKKGSNLESNAKESQTERKPTFSLELVNGWHLI 64
Query 80 CSADGKSYWFHSATKRSVWNTPKEVEEMLKSLKE 113
DG +F+ +K + +E + +SL E
Sbjct 65 IYNDGTKLYFNDDSKEFKNDISQEDDSRCRSLIE 98
> hsa:51741 WWOX, D16S432E, FOR, FRA16D, HHCMA56, PRO0128, SDR41C1,
WOX1; WW domain containing oxidoreductase (EC:1.1.1.-)
Length=414
Score = 35.4 bits (80), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 31/152 (20%), Positives = 64/152 (42%), Gaps = 20/152 (13%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQIL--LKTGWQEFCSADGKSYWFH 90
GW E +K+G YY N +++QW P +T + ++ L GW++ +G+ ++
Sbjct 21 GWEERTTKDGWVYYANHTEEKTQWEHP---KTGKRKRVAGDLPYGWEQETDENGQVFFVD 77
Query 91 SATKRSVWNTPKEVEEMLKSLKEEREDWPQFKSKEEAKTFLVKLFELKKFPPR------- 143
KR+ + P+ L ++ P + + + T +++ + + F +
Sbjct 78 HINKRTTYLDPR-----LAFTVDDNPTKPTTRQRYDGSTTAMEILQGRDFTGKVVVVTGA 132
Query 144 ---ISWENASKILESRPQVVLLLHFDARRAES 172
I +E A V+L AR +E+
Sbjct 133 NSGIGFETAKSFALHGAHVILACRNMARASEA 164
> tgo:TGME49_121360 RNA recognition motif domain-containing protein
Length=274
Score = 35.0 bits (79), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 34 WTEHISKEGRKYYYNTLTQESQWGKPLSLQTP 65
W E+ + EG YYYNT T +QW KP P
Sbjct 110 WQEYFTPEGYAYYYNTSTGVTQWEKPEDFDKP 141
> dre:559634 nedd4l; neural precursor cell expressed, developmentally
down-regulated 4-like; K13305 E3 ubiquitin-protein ligase
NEDD4-like [EC:6.3.2.19]
Length=994
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 36/80 (45%), Gaps = 11/80 (13%)
Query 33 GWTEHISKEGRKYYYNTLTQESQWGKPLSLQTPEEAQI----------LLKTGWQEFCSA 82
GW I+ GR ++ + ++ + W P L+ P + L GW+E A
Sbjct 521 GWEMRIAPNGRPFFIDHNSRTTTWEDP-RLKYPVHMRTKASLDPGDLGPLPPGWEERVHA 579
Query 83 DGKSYWFHSATKRSVWNTPK 102
DG++++ K++ W P+
Sbjct 580 DGRTFYIDHNNKKTQWEDPR 599
Lambda K H
0.313 0.128 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4406352944
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40