bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2552_orf1
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
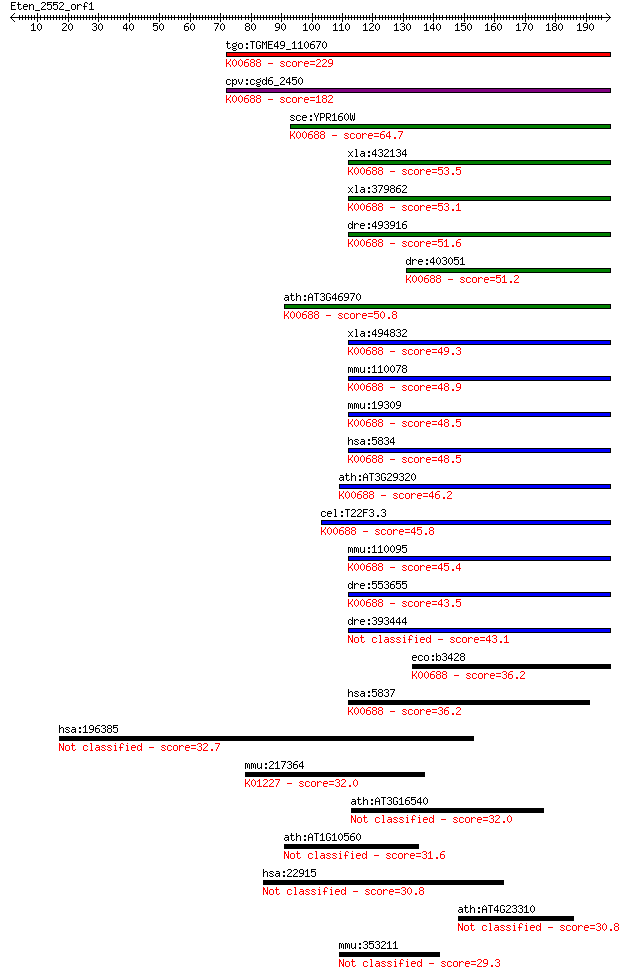
tgo:TGME49_110670 glycogen phosphorylase family protein, putat... 229 5e-60
cpv:cgd6_2450 glycogen phosphorylase ; K00688 starch phosphory... 182 6e-46
sce:YPR160W GPH1; Gph1p (EC:2.4.1.1); K00688 starch phosphoryl... 64.7 2e-10
xla:432134 hypothetical protein MGC80198; K00688 starch phosph... 53.5 5e-07
xla:379862 pygm, MGC53328, pygb; phosphorylase, glycogen, musc... 53.1 5e-07
dre:493916 pygl, zgc:66314; phosphorylase, glycogen; liver (He... 51.6 2e-06
dre:403051 pygb; phosphorylase, glycogen; brain (EC:2.4.1.1); ... 51.2 2e-06
ath:AT3G46970 PHS2; PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosp... 50.8 3e-06
xla:494832 pygl; phosphorylase, glycogen, liver (EC:2.4.1.1); ... 49.3 8e-06
mmu:110078 Pygb, MGC36329; brain glycogen phosphorylase (EC:2.... 48.9 1e-05
mmu:19309 Pygm, AI115133, PG; muscle glycogen phosphorylase (E... 48.5 1e-05
hsa:5834 PYGB, MGC9213; phosphorylase, glycogen; brain (EC:2.4... 48.5 1e-05
ath:AT3G29320 glucan phosphorylase, putative; K00688 starch ph... 46.2 8e-05
cel:T22F3.3 hypothetical protein; K00688 starch phosphorylase ... 45.8 1e-04
mmu:110095 Pygl; liver glycogen phosphorylase (EC:2.4.1.1); K0... 45.4 1e-04
dre:553655 pygma, MGC110706, im:7150327, zgc:110706; phosphory... 43.5 4e-04
dre:393444 pygmb, MGC63642, zgc:63642; phosphorylase, glycogen... 43.1 6e-04
eco:b3428 glgP, ECK3414, glgY, JW3391; glycogen phosphorylase ... 36.2 0.076
hsa:5837 PYGM; phosphorylase, glycogen, muscle (EC:2.4.1.1); K... 36.2 0.076
hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dyn... 32.7 0.82
mmu:217364 Engase, C130099A03, D230014K01Rik; endo-beta-N-acet... 32.0 1.4
ath:AT3G16540 DegP11; DegP11 (DegP protease 11); catalytic/ pr... 32.0 1.5
ath:AT1G10560 PUB18; PUB18 (PLANT U-BOX 18); ubiquitin-protein... 31.6 1.9
hsa:22915 MMRN1, ECM, EMILIN4, GPIa*, MMRN; multimerin 1 30.8
ath:AT4G23310 receptor-like protein kinase, putative 30.8 3.4
mmu:353211 Prune2, 6330414G02Rik, A230083H22Rik, A330102H22Rik... 29.3 9.9
> tgo:TGME49_110670 glycogen phosphorylase family protein, putative
(EC:2.4.1.1); K00688 starch phosphorylase [EC:2.4.1.1]
Length=925
Score = 229 bits (584), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 106/126 (84%), Positives = 117/126 (92%), Gaps = 1/126 (0%)
Query 72 EDVFTSDTNQHWEMKRKASFSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPS 131
E+VF +D++ HWEM+RKASFSKLTGAVPR IPGMYN EDPHAD+KKEKLWKLMETY+ S
Sbjct 7 ENVFANDSSYHWEMRRKASFSKLTGAVPRAIPGMYNE-EDPHADEKKEKLWKLMETYISS 65
Query 132 DVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRA 191
D+HSIQRSIVNHVEYT A+TRFN DPESCYRA+AFSVRDRLIETLNDTNA+FHEKD KRA
Sbjct 66 DIHSIQRSIVNHVEYTCARTRFNCDPESCYRASAFSVRDRLIETLNDTNAYFHEKDCKRA 125
Query 192 YYLSLE 197
YYLSLE
Sbjct 126 YYLSLE 131
> cpv:cgd6_2450 glycogen phosphorylase ; K00688 starch phosphorylase
[EC:2.4.1.1]
Length=901
Score = 182 bits (462), Expect = 6e-46, Method: Composition-based stats.
Identities = 86/126 (68%), Positives = 103/126 (81%), Gaps = 1/126 (0%)
Query 72 EDVFTSDTNQHWEMKRKASFSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPS 131
+ VFT D N ++EM+RKASFSKLTGAVPR + GMY DP AD ++EKLW LME+YLP+
Sbjct 3 DSVFTRD-NYNFEMRRKASFSKLTGAVPRGMTGMYLDDFDPTADKRREKLWYLMESYLPT 61
Query 132 DVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRA 191
D+ SIQRSIVNHVEYTLA+TRFN D + YRA A+S+RDRLIE LNDTN +F+E+D KR
Sbjct 62 DIESIQRSIVNHVEYTLARTRFNFDDNAAYRATAYSIRDRLIENLNDTNEYFNERDCKRC 121
Query 192 YYLSLE 197
YYLSLE
Sbjct 122 YYLSLE 127
> sce:YPR160W GPH1; Gph1p (EC:2.4.1.1); K00688 starch phosphorylase
[EC:2.4.1.1]
Length=902
Score = 64.7 bits (156), Expect = 2e-10, Method: Composition-based stats.
Identities = 37/105 (35%), Positives = 53/105 (50%), Gaps = 5/105 (4%)
Query 93 KLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTR 152
+LTG +P+ I + D K LW + + Q ++HVE TLA++
Sbjct 29 RLTGFLPQEIKSI-----DTMIPLKSRALWNKHQVKKFNKAEDFQDRFIDHVETTLARSL 83
Query 153 FNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
+N D + Y AA+ S+RD L+ N T F +D KR YYLSLE
Sbjct 84 YNCDDMAAYEAASMSIRDNLVIDWNKTQQKFTTRDPKRVYYLSLE 128
> xla:432134 hypothetical protein MGC80198; K00688 starch phosphorylase
[EC:2.4.1.1]
Length=843
Score = 53.5 bits (127), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D K+K + DV +++S H+ +TL K R P Y A A +VRD
Sbjct 4 PLSDSDKKKQISVRGIAGLGDVSEVRKSFNRHLHFTLVKDRNVSTPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T +++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQYYYEKDPKRIYYLSLE 89
> xla:379862 pygm, MGC53328, pygb; phosphorylase, glycogen, muscle
(EC:2.4.1.1); K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/86 (37%), Positives = 45/86 (52%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D K+K + DV +++S H+ +TL K R P Y A A +VRD
Sbjct 4 PLSDSDKKKQISVRGIAGLGDVSEVRKSFNRHLHFTLVKDRNVSTPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T +++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQYYYEKDPKRIYYLSLE 89
> dre:493916 pygl, zgc:66314; phosphorylase, glycogen; liver (Hers
disease, glycogen storage disease type VI) (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=967
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
PH D +K K + +V +++ H+ +TL K R P Y A + +VRD
Sbjct 4 PHTDQEKRKQISIRGIVGVENVAELKKGFNRHLHFTLVKDRNVATPRDYYFALSHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T F +E D KR YYLSLE
Sbjct 64 LVGRWIRTQQFCYEADPKRVYYLSLE 89
> dre:403051 pygb; phosphorylase, glycogen; brain (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/67 (41%), Positives = 38/67 (56%), Gaps = 0/67 (0%)
Query 131 SDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKR 190
DV I++S H+ +TL K R P Y A A +VRD L+ T +++EKD KR
Sbjct 23 GDVAEIKKSFNRHLHFTLVKDRNVATPRDYYFALAHTVRDHLVGRWIRTQQYYYEKDPKR 82
Query 191 AYYLSLE 197
+YLSLE
Sbjct 83 IHYLSLE 89
> ath:AT3G46970 PHS2; PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosphorylase/
transferase, transferring glycosyl groups; K00688
starch phosphorylase [EC:2.4.1.1]
Length=841
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/107 (29%), Positives = 48/107 (44%), Gaps = 16/107 (14%)
Query 91 FSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAK 150
+ G ++P + +P ADD E I +IV H +Y+
Sbjct 1 MANANGKAATSLPEKISAKANPEADDATE----------------IAGNIVYHAKYSPHF 44
Query 151 TRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
+ PE A A S+RDRLI+ N+T F++ D K+ YYLS+E
Sbjct 45 SPLKFGPEQALYATAESLRDRLIQLWNETYVHFNKVDPKQTYYLSME 91
> xla:494832 pygl; phosphorylase, glycogen, liver (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=855
Score = 49.3 bits (116), Expect = 8e-06, Method: Composition-based stats.
Identities = 29/86 (33%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K K + +V +++ H+ +TL K R Y A A +VRD
Sbjct 4 PLTDQEKRKQISIRGIVGVENVAELKKGFNRHLHFTLVKDRNVATIRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T +++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQYYYEKDPKRTYYLSLE 89
> mmu:110078 Pygb, MGC36329; brain glycogen phosphorylase (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +++K + DV +++S H+ +TL K R P + A A +VRD
Sbjct 4 PLTDSERQKQISVRGIAGLGDVAEVRKSFNRHLHFTLVKDRNVATPRDYFFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++E+D KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYERDPKRIYYLSLE 89
> mmu:19309 Pygm, AI115133, PG; muscle glycogen phosphorylase
(EC:2.4.1.1); K00688 starch phosphorylase [EC:2.4.1.1]
Length=842
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D K K + +V ++++ H+ +TL K R P Y A A +VRD
Sbjct 4 PLSDQDKRKQISVRGLAGVENVSELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYEKDPKRIYYLSLE 89
> hsa:5834 PYGB, MGC9213; phosphorylase, glycogen; brain (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=843
Score = 48.5 bits (114), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 30/86 (34%), Positives = 43/86 (50%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K K + DV +++S H+ +TL K R P + A A +VRD
Sbjct 4 PLTDSEKRKQISVRGLAGLGDVAEVRKSFNRHLHFTLVKDRNVATPRDYFFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++E+D KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYERDPKRIYYLSLE 89
> ath:AT3G29320 glucan phosphorylase, putative; K00688 starch
phosphorylase [EC:2.4.1.1]
Length=962
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 34/91 (37%), Positives = 50/91 (54%), Gaps = 5/91 (5%)
Query 109 VEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYT--LAKTRFNLDPESCYRAAAF 166
V D D ++E M + P D S+ SI H E+T + +F L P++ + A A
Sbjct 70 VTDAVLDSEQEVFISSMNPFAP-DAASVASSIKYHAEFTPLFSPEKFEL-PKAFF-ATAQ 126
Query 167 SVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
SVRD LI N T +++ +VK+AYYLS+E
Sbjct 127 SVRDALIMNWNATYEYYNRVNVKQAYYLSME 157
> cel:T22F3.3 hypothetical protein; K00688 starch phosphorylase
[EC:2.4.1.1]
Length=882
Score = 45.8 bits (107), Expect = 1e-04, Method: Composition-based stats.
Identities = 26/95 (27%), Positives = 50/95 (52%), Gaps = 3/95 (3%)
Query 103 PGMYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYR 162
P +++++ + H K+ + + + +V +I+++ H+ +++ K R Y
Sbjct 32 PKLFSMITNDHDRRKQISVRGIAQV---ENVSNIKKAFNRHLHFSIIKDRNVATDRDYYF 88
Query 163 AAAFSVRDRLIETLNDTNAFFHEKDVKRAYYLSLE 197
A A +VRD L+ T +++KD KR YYLSLE
Sbjct 89 ALANTVRDHLVSRWIRTQQHYYDKDPKRVYYLSLE 123
> mmu:110095 Pygl; liver glycogen phosphorylase (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=850
Score = 45.4 bits (106), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K + + +V +++ H+ +TL K R P Y A A +VRD
Sbjct 4 PLTDQEKRRQISIRGIVGVENVAELKKGFNRHLHFTLVKDRNVATPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T +++K KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYDKCPKRVYYLSLE 89
> dre:553655 pygma, MGC110706, im:7150327, zgc:110706; phosphorylase,
glycogen (muscle) A (EC:2.4.1.1); K00688 starch phosphorylase
[EC:2.4.1.1]
Length=842
Score = 43.5 bits (101), Expect = 4e-04, Method: Composition-based stats.
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D ++K + +V ++ + H+ +TL K R Y A A +VRD
Sbjct 4 PLSDHDRKKQISVRGLAGVENVADLKTNFNRHLHFTLVKDRNVSTKRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++EKD KR YY+SLE
Sbjct 64 LVGRWIRTQQSYYEKDPKRVYYISLE 89
> dre:393444 pygmb, MGC63642, zgc:63642; phosphorylase, glycogen
(muscle) b
Length=315
Score = 43.1 bits (100), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 41/86 (47%), Gaps = 0/86 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P D +K K + +V ++ + H+ +TL K R Y A A +VRD
Sbjct 4 PLTDQEKRKQISVRGLAGVENVADLKTNFNRHLHFTLVKDRNVATKRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKRAYYLSLE 197
L+ T ++EKD KR YYLSLE
Sbjct 64 LVGRWIRTQQHYYEKDPKRVYYLSLE 89
> eco:b3428 glgP, ECK3414, glgY, JW3391; glycogen phosphorylase
(EC:2.4.1.1); K00688 starch phosphorylase [EC:2.4.1.1]
Length=815
Score = 36.2 bits (82), Expect = 0.076, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 133 VHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDRLIETLNDTNAFFHEKDVKRAY 192
V +++ SI + +T+ K + A F+VRDRL+E +N ++ ++ Y
Sbjct 14 VEALKHSIAYKLMFTIGKDPVVANKHEWLNATLFAVRDRLVERWLRSNRAQLSQETRQVY 73
Query 193 YLSLE 197
YLS+E
Sbjct 74 YLSME 78
> hsa:5837 PYGM; phosphorylase, glycogen, muscle (EC:2.4.1.1);
K00688 starch phosphorylase [EC:2.4.1.1]
Length=754
Score = 36.2 bits (82), Expect = 0.076, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 38/79 (48%), Gaps = 0/79 (0%)
Query 112 PHADDKKEKLWKLMETYLPSDVHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVRDR 171
P +D +K K + +V ++++ H+ +TL K R P Y A A +VRD
Sbjct 4 PLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHTVRDH 63
Query 172 LIETLNDTNAFFHEKDVKR 190
L+ T ++EKD K+
Sbjct 64 LVGRWIRTQQHYYEKDPKK 82
> hsa:196385 DNAH10, FLJ38262, FLJ43486, FLJ43808, KIAA2017; dynein,
axonemal, heavy chain 10
Length=4471
Score = 32.7 bits (73), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 57/142 (40%), Gaps = 20/142 (14%)
Query 17 LPRPLLRSVSVL--LLPRLEEGAVRLRRTRSAYFLAFFLLFLYLRLLTD----FVKMATG 70
+P+ L V+ L +L L EG + YFL L LL D F +
Sbjct 2309 VPQTDLNMVTQLAKMLDALLEGEIEDLDLLECYFLEALYCSLGASLLEDGRMKFDEYIKR 2368
Query 71 VEDVFTSDTNQHWEMKRKASFSKLTGAVPRTIPGMYNVVEDPHADDKKEKLWKLMETYLP 130
+ + T DT W A P +PG + D H D+K+ + W +P
Sbjct 2369 LASLSTVDTEGVW-------------ANPGELPGQLPTLYDFHFDNKRNQ-WVPWSKLVP 2414
Query 131 SDVHSIQRSIVNHVEYTLAKTR 152
+H+ +R +N + +T+ TR
Sbjct 2415 EYIHAPERKFINILVHTVDTTR 2436
> mmu:217364 Engase, C130099A03, D230014K01Rik; endo-beta-N-acetylglucosaminidase
(EC:3.2.1.96); K01227 mannosyl-glycoprotein
endo-beta-N-acetylglucosaminidase [EC:3.2.1.96]
Length=734
Score = 32.0 bits (71), Expect = 1.4, Method: Composition-based stats.
Identities = 21/60 (35%), Positives = 31/60 (51%), Gaps = 8/60 (13%)
Query 78 DTNQHWEMKRKASFSKLTGAVPRTIPG-MYNVVEDPHADDKKEKLWKLMETYLPSDVHSI 136
DT++ E+ RK FS A PG +Y +E ++K W L+E +LP+ HSI
Sbjct 335 DTDKSLELIRKHGFSAALFA-----PGWVYECLEKSDFFQNQDKFWSLLERFLPT--HSI 387
> ath:AT3G16540 DegP11; DegP11 (DegP protease 11); catalytic/
protein binding / serine-type endopeptidase/ serine-type peptidase
Length=555
Score = 32.0 bits (71), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 113 HADDKKEKLWKLMETYLPSD---VHSIQRSIVNHVEYTLAKTRFNLDPESCYRAAAFSVR 169
H D+KK + WK +E P D + S+ + N EY+ +K LD +S R F++
Sbjct 83 HEDEKKLERWKKIEESHPLDELVLDSVVKVFSNSTEYSKSKPWKTLDQKSS-RGTGFAIA 141
Query 170 DRLIET 175
R I T
Sbjct 142 GRKILT 147
> ath:AT1G10560 PUB18; PUB18 (PLANT U-BOX 18); ubiquitin-protein
ligase
Length=697
Score = 31.6 bits (70), Expect = 1.9, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query 91 FSKLTGAVPRTIPGMYNVVE-DPHADD-KKEKLWKLMETYLPSDVH 134
+S+L G P IPG+ N+V+ D + D K+ L +M + SD H
Sbjct 504 YSRLIGENPDAIPGLMNIVKGDDYGDSAKRSALLAVMGLLMQSDNH 549
> hsa:22915 MMRN1, ECM, EMILIN4, GPIa*, MMRN; multimerin 1
Length=1228
Score = 30.8 bits (68), Expect = 2.8, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 19/87 (21%)
Query 84 EMKRKASFSKLTGAVPRTIPG-MYNVVEDPHADDKKEKLWKLMETYLPSDVHSIQRSIVN 142
++K +A+ S LT + R++PG + NVV K +K K LP ++++++ VN
Sbjct 972 QIKTQAALSNLTCCIDRSLPGSLANVV-------KSQKQVK----SLPKKINALKKPTVN 1020
Query 143 HVEYTLAKTRFNLD----PE---SCYR 162
+ +T+ N D PE SC R
Sbjct 1021 LTTVLIGRTQRNTDNIIYPEEYSSCSR 1047
> ath:AT4G23310 receptor-like protein kinase, putative
Length=830
Score = 30.8 bits (68), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 21/40 (52%), Gaps = 2/40 (5%)
Query 148 LAKTRFNLDPESCYRAAAFSVRDRLIETLN--DTNAFFHE 185
L R +L PE C+ AF+V+D LI N D F+ E
Sbjct 84 LFDCRGDLPPEVCHNCVAFAVKDTLIRCPNERDVTLFYDE 123
> mmu:353211 Prune2, 6330414G02Rik, A230083H22Rik, A330102H22Rik,
Bmcc1, KIAA0367, mKIAA0367; prune homolog 2 (Drosophila)
Length=3084
Score = 29.3 bits (64), Expect = 9.9, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 18/33 (54%), Gaps = 0/33 (0%)
Query 109 VEDPHADDKKEKLWKLMETYLPSDVHSIQRSIV 141
V D H D +EKLW + L SD + + + ++
Sbjct 1878 VLDSHGDKSQEKLWNIQPKQLDSDANQLSQLVI 1910
Lambda K H
0.325 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5802328440
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40