bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2551_orf4
Length=277
Score E
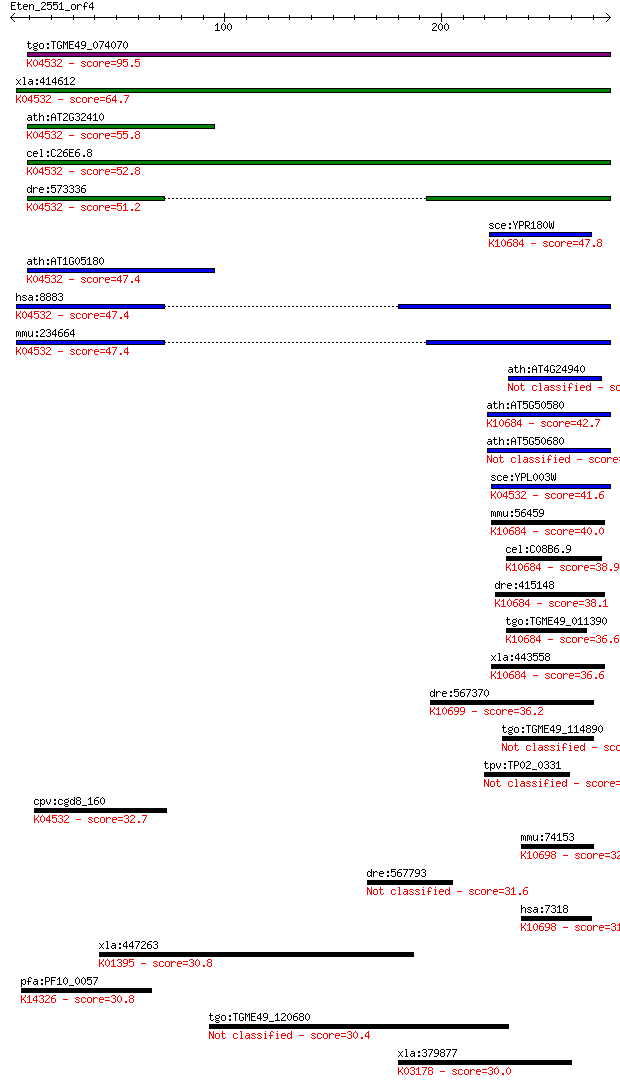
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_074070 thiF family domain-containing protein ; K045... 95.5 2e-19
xla:414612 nae1, MGC81483; NEDD8 activating enzyme E1 subunit ... 64.7 3e-10
ath:AT2G32410 AXL; AXL (AXR1-LIKE); binding / catalytic; K0453... 55.8 2e-07
cel:C26E6.8 ula-1; yeast ULA (ubiquitin activating) homolog fa... 52.8 1e-06
dre:573336 nae1, MGC171400, MGC66148, appbp1, zgc:66148; nedd8... 51.2 4e-06
sce:YPR180W AOS1, RHC31; Aos1p; K10684 ubiquitin-like 1-activa... 47.8 5e-05
ath:AT1G05180 AXR1; AXR1 (AUXIN RESISTANT 1); small protein ac... 47.4 5e-05
hsa:8883 NAE1, A-116A10.1, APPBP1, ula-1; NEDD8 activating enz... 47.4 5e-05
mmu:234664 Nae1, 59kDa, Appbp1, MGC29435, MGC36437, MGC36630; ... 47.4 6e-05
ath:AT4G24940 SAE1A; SAE1A (SUMO-ACTIVATING ENZYME 1A); SUMO a... 42.7 0.001
ath:AT5G50580 SAE1B; SAE1B (SUMO-ACTIVATING ENZYME 1B); SUMO a... 42.7 0.002
ath:AT5G50680 SAE1B; SAE1B (SUMO ACTIVATING ENZYME 1B); SUMO a... 42.7 0.002
sce:YPL003W ULA1, ENR2; Protein that acts together with Uba3p ... 41.6 0.003
mmu:56459 Sae1, 2400010M20Rik, 2610044L12Rik, AL033372, AOS1, ... 40.0 0.010
cel:C08B6.9 aos-1; Activator Of Sumo (yeast AOS homolog) famil... 38.9 0.023
dre:415148 sae1, sae2a, uble1a, wu:fa28b04, zgc:86633; SUMO1 a... 38.1 0.038
tgo:TGME49_011390 hypothetical protein ; K10684 ubiquitin-like... 36.6 0.094
xla:443558 sae1, Aos1p, MGC78909, Sua1p, aos; SUMO1 activating... 36.6 0.11
dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquit... 36.2 0.15
tgo:TGME49_114890 ubiquitin-activating enzyme E1, putative 34.3 0.45
tpv:TP02_0331 ubiquitin activating enzyme, putatuve 34.3 0.48
cpv:cgd8_160 ubiquitin activating enzyme E1 ; K04532 amyloid b... 32.7 1.5
mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier ... 32.3 2.0
dre:567793 transmembrane channel-like 3-like 31.6 3.0
hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-lik... 31.6 3.0
xla:447263 MGC86332 protein; K01395 meprin A, alpha [EC:3.4.24... 30.8 5.3
pfa:PF10_0057 regulator of nonsense transcripts, putative; K14... 30.8 5.3
tgo:TGME49_120680 hypothetical protein 30.4 7.7
xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1, p... 30.0 8.7
> tgo:TGME49_074070 thiF family domain-containing protein ; K04532
amyloid beta precursor protein binding protein 1
Length=779
Score = 95.5 bits (236), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 89/345 (25%), Positives = 142/345 (41%), Gaps = 104/345 (30%)
Query 9 DFAADTKSYLELQKIYNEKAERDERRLR-------------ELLGQLDVHP--------- 46
D +DT+S++ LQ++Y+++AE D ++ E GQ H
Sbjct 463 DMTSDTQSFIRLQQVYSKRAEGDCNAIKASVAMIQEDSITLEGDGQGRRHGGVTNSQRSC 522
Query 47 SAKLNVSQE-----------------------EIKCFCKNAVFFKVTQYPKITGAFLGST 83
S L SQ +++ FC+NA KV +Y I F T
Sbjct 523 SPPLRCSQLSSREGSVGSAFRRRDSERSGASLDVEKFCRNAYNLKVIRYRSIGEEFNPLT 582
Query 84 SVELLAAAAREMQQAVPEELKLSPNADPDLFADPNFSVYEENGFLHLPWYAAALACRAAA 143
REM L++ + + G LPWY A AC A
Sbjct 583 -------VNREMYGEAVAGLRMEES---------------DEGIPLLPWYLALWACHRFA 620
Query 144 AKLGRFPG----------SHPKPSAAAATAATGEAAAAATGG------------------ 175
A+ G FPG S + S + AT + T G
Sbjct 621 ARNGFFPGVRAVGWLDANSKQEMSLSKATTTHWDVDKNETTGRCLNPQYQCESEEGEQHR 680
Query 176 ---AAAARSVEEAIKQQQELVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSE 232
+ AR E ++ E ++++ +L E+ + L ++++Q+V++ G+E
Sbjct 681 TQVSVTARPPEMSLDAAIEGLKREVNVLLSEINVPD------LTVDERIIRQMVAFGGAE 734
Query 233 FPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
TAA+VG VAAQEA+KL+ ++FEPINNTF+WNG++R+ ++L
Sbjct 735 IHTTAAIVGGVAAQEAVKLICRQFEPINNTFIWNGIERKAEVLEL 779
> xla:414612 nae1, MGC81483; NEDD8 activating enzyme E1 subunit
1; K04532 amyloid beta precursor protein binding protein 1
Length=533
Score = 64.7 bits (156), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 65/278 (23%), Positives = 107/278 (38%), Gaps = 73/278 (26%)
Query 4 RRLTADFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKC 59
R D AD+ +++LQ IY EKA++D E + +LL + P + +S+ +I+
Sbjct 325 RGTIPDMIADSDKFIKLQNIYREKAKKDASAVESCVSKLLQSVGRPPES---ISERDIRL 381
Query 60 FCKNAVFFKVTQYPKITGAFLGSTSVELLAAAAREMQQAVPEELKLSPNADPDLFADPNF 119
FC+N F +V + +++ EE L D+ +
Sbjct 382 FCRNCAFLRVVRC------------------------RSLEEEYGLDTAKKDDIVS---- 413
Query 120 SVYEENGFLHLPWYAAALACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAA 179
EN + Y A +LGR+PG
Sbjct 414 --LMENPDNEIVLYLMLRAVDRFQKQLGRYPG---------------------------- 443
Query 180 RSVEEAIKQQQELVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAAL 239
I Q +E D + L + + G L + +Q+ Y +E A+
Sbjct 444 ------IYNYQ--IESDIGKLKSCLNGLLQEYGLSLTVKDEYIQEFCRYGAAEPHTIASF 495
Query 240 VGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
+G AAQEAIK++ K+F NNTF++N + + T QL
Sbjct 496 LGGAAAQEAIKIITKQFVIFNNTFIYNAMLQTSATFQL 533
> ath:AT2G32410 AXL; AXL (AXR1-LIKE); binding / catalytic; K04532
amyloid beta precursor protein binding protein 1
Length=417
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 7/90 (7%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D + T+ Y+ LQKIY+ KAE D E+R++ +L ++ PS+ +S+ IK FCKNA
Sbjct 319 DMISSTEHYINLQKIYHSKAEADFLSMEQRVKSILVKVGQDPSS---ISKPTIKSFCKNA 375
Query 65 VFFKVTQYPKITGAFLGSTSVELLAAAARE 94
KV +Y I F ++ EL A E
Sbjct 376 RKLKVCRYRTIEDEFKSPSTTELHKYLADE 405
> cel:C26E6.8 ula-1; yeast ULA (ubiquitin activating) homolog
family member (ula-1); K04532 amyloid beta precursor protein
binding protein 1
Length=541
Score = 52.8 bits (125), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 61/269 (22%), Positives = 104/269 (38%), Gaps = 53/269 (19%)
Query 9 DFAADTKSYLELQKIYNEKAERDERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNAVFFK 68
D +D+ Y L +++EKA D + + L +++ +S + FCKNA +
Sbjct 326 DMTSDSSRYTRLATLFHEKALSDAQEVLRLTREVEKERGVGDVISDDVCYRFCKNADRIR 385
Query 69 VTQYPKITGAFLGSTSVELLAAAAREMQQAVPEELKLSPNADPDLFADPNFSVYEENGFL 128
V QY + E +A+ E+++ S N D + N V E
Sbjct 386 V-QYGDVLDY--------------NEETKAIVEKIRES-NIDEET---RNQKVDEAT--- 423
Query 129 HLPWYAAALACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAARSVEEAIKQ 188
W A + GR+PG++ P + A + EA+K
Sbjct 424 ---WMLLMRAVGRFQKEKGRYPGTNGVPVSIDAQDLKKRVEVL----------IREALKD 470
Query 189 QQELVEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEA 248
+Q+ + + E+ CR AA E ++ VG +AAQE
Sbjct 471 EQDFTSISNKVTDTAIAEI-----CRFGAA-------------ELHVISSYVGGIAAQEI 512
Query 249 IKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
IKL ++ PI+NTF+++G + T +
Sbjct 513 IKLATNQYVPIDNTFIFDGHTQESATFKF 541
> dre:573336 nae1, MGC171400, MGC66148, appbp1, zgc:66148; nedd8
activating enzyme E1 subunit 1; K04532 amyloid beta precursor
protein binding protein 1
Length=533
Score = 51.2 bits (121), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 43/85 (50%), Gaps = 0/85 (0%)
Query 193 VEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEAIKLL 252
VE+D + L + + + + + + Y +E AA +G AAQEAIK++
Sbjct 449 VEEDINKLKLCVNSLLQEYSLNVNVKDDYIHEFCRYGAAEPHTVAAFLGGSAAQEAIKII 508
Query 253 NKKFEPINNTFLWNGVQRRGLTMQL 277
+F P NNTFL+N + + T QL
Sbjct 509 THQFVPFNNTFLYNAMSQTSATFQL 533
Score = 46.6 bits (109), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 39/67 (58%), Gaps = 7/67 (10%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D AD+ +++LQ +Y +KA RD + + LL + P + +S++EIK FCKNA
Sbjct 330 DMIADSDKFIKLQNVYRDKAMRDAAVVSKHVEMLLQSVGKTPES---ISEQEIKLFCKNA 386
Query 65 VFFKVTQ 71
F +V +
Sbjct 387 AFLRVVR 393
> sce:YPR180W AOS1, RHC31; Aos1p; K10684 ubiquitin-like 1-activating
enzyme E1 A [EC:6.3.2.19]
Length=347
Score = 47.8 bits (112), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 30/47 (63%), Gaps = 0/47 (0%)
Query 222 VQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGV 268
+QQ + +G EF AA++G AQ+ I +L K+ P+NN +++G+
Sbjct 292 IQQFIKQKGIEFAPVAAIIGGAVAQDVINILGKRLSPLNNFIVFDGI 338
> ath:AT1G05180 AXR1; AXR1 (AUXIN RESISTANT 1); small protein
activating enzyme; K04532 amyloid beta precursor protein binding
protein 1
Length=422
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 7/90 (7%)
Query 9 DFAADTKSYLELQKIYNEKAERD----ERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNA 64
D + T+ Y+ LQKIY KAE D E R++ +L ++ PS+ + + IK FCKNA
Sbjct 232 DMTSSTEHYINLQKIYLAKAEADFLVIEERVKNILKKIGRDPSS---IPKPTIKSFCKNA 288
Query 65 VFFKVTQYPKITGAFLGSTSVELLAAAARE 94
K+ +Y + F + E+ A E
Sbjct 289 RKLKLCRYRMVEDEFRNPSVTEIQKYLADE 318
> hsa:8883 NAE1, A-116A10.1, APPBP1, ula-1; NEDD8 activating enzyme
E1 subunit 1; K04532 amyloid beta precursor protein binding
protein 1
Length=528
Score = 47.4 bits (111), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 49/105 (46%), Gaps = 7/105 (6%)
Query 180 RSVEEAIKQQQEL-------VEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSE 232
R+V+ KQQ VE+D + L + G + V + Y +E
Sbjct 424 RAVDRFHKQQGRYPGVSNYQVEEDIGKLKSCLTGFLQEYGLSVMVKDDYVHEFCRYGAAE 483
Query 233 FPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
AA +G AAQE IK++ K+F NNT++++G+ + T QL
Sbjct 484 PHTIAAFLGGAAAQEVIKIITKQFVIFNNTYIYSGMSQTSATFQL 528
Score = 41.2 bits (95), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query 4 RRLTADFAADTKSYLELQKIYNEKAERDE----RRLRELLGQLDVHPSAKLNVSQEEIKC 59
R D AD+ Y++LQ +Y EKA++D + +LL + P + +S++E+K
Sbjct 320 RGTIPDMIADSGKYIKLQNVYREKAKKDAAAVGNHVAKLLQSIGQAPES---ISEKELKL 376
Query 60 FCKNAVFFKVTQ 71
C N+ F +V +
Sbjct 377 LCSNSAFLRVVR 388
> mmu:234664 Nae1, 59kDa, Appbp1, MGC29435, MGC36437, MGC36630;
NEDD8 activating enzyme E1 subunit 1; K04532 amyloid beta
precursor protein binding protein 1
Length=534
Score = 47.4 bits (111), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 42/85 (49%), Gaps = 0/85 (0%)
Query 193 VEKDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVSYRGSEFPATAALVGAVAAQEAIKLL 252
VE+D + L + G + V + Y +E AA +G AAQE IK++
Sbjct 450 VEEDIGKLKSCLTGFLQEYGLSVMVKDDYVHEFCRYGAAEPHTIAAFLGGAAAQEVIKII 509
Query 253 NKKFEPINNTFLWNGVQRRGLTMQL 277
K+F NNT++++G+ + T QL
Sbjct 510 TKQFVIFNNTYIYSGMSQTSATFQL 534
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 39/72 (54%), Gaps = 7/72 (9%)
Query 4 RRLTADFAADTKSYLELQKIYNEKAERDE----RRLRELLGQLDVHPSAKLNVSQEEIKC 59
R D AD+ Y++LQ +Y EKA++D + +LL + P + +S++E+K
Sbjct 326 RGTIPDMIADSNKYIKLQNVYREKAKKDAAAVGNHVAKLLQSVGQAPES---ISEKELKL 382
Query 60 FCKNAVFFKVTQ 71
C N+ F +V +
Sbjct 383 LCSNSAFLRVVR 394
> ath:AT4G24940 SAE1A; SAE1A (SUMO-ACTIVATING ENZYME 1A); SUMO
activating enzyme
Length=322
Score = 42.7 bits (99), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 28/43 (65%), Gaps = 0/43 (0%)
Query 231 SEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGL 273
+EFP A+VG + AQE IK ++ K +P+ N F ++G +G+
Sbjct 270 TEFPPVCAIVGGILAQEVIKAVSGKGDPLKNFFYYDGEDGKGV 312
> ath:AT5G50580 SAE1B; SAE1B (SUMO-ACTIVATING ENZYME 1B); SUMO
activating enzyme; K10684 ubiquitin-like 1-activating enzyme
E1 A [EC:6.3.2.19]
Length=320
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 221 VVQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
++++LVS +EFP A++G + QE IK+++ K EP+ N F ++ +G+ L
Sbjct 261 LLERLVS-NNTEFPPACAIIGGILGQEVIKVISGKGEPLKNFFYFDAEDGKGVIEDL 316
> ath:AT5G50680 SAE1B; SAE1B (SUMO ACTIVATING ENZYME 1B); SUMO
activating enzyme
Length=318
Score = 42.7 bits (99), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query 221 VVQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
++++LVS +EFP A++G + QE IK+++ K EP+ N F ++ +G+ L
Sbjct 259 LLERLVS-NNTEFPPACAIIGGILGQEVIKVISGKGEPLKNFFYFDAEDGKGVIEDL 314
> sce:YPL003W ULA1, ENR2; Protein that acts together with Uba3p
to activate Rub1p before its conjugation to proteins (neddylation),
which may play a role in protein degradation; K04532
amyloid beta precursor protein binding protein 1
Length=462
Score = 41.6 bits (96), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 0/55 (0%)
Query 223 QQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLTMQL 277
+ + R + + A G QEAIKL+ + PI+N FL+NG+ T ++
Sbjct 408 DEFIGLRVDDNYSVMAFFGGAVVQEAIKLITHHYVPIDNLFLYNGINNSSATYKI 462
> mmu:56459 Sae1, 2400010M20Rik, 2610044L12Rik, AL033372, AOS1,
AW743391, D7Ertd177e, HSPC140, Sua1, Uble1a; SUMO1 activating
enzyme subunit 1; K10684 ubiquitin-like 1-activating enzyme
E1 A [EC:6.3.2.19]
Length=350
Score = 40.0 bits (92), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 29/52 (55%), Gaps = 0/52 (0%)
Query 223 QQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLT 274
V Y SE A+VG + AQE +K L+++ P NN F ++G++ G+
Sbjct 293 DDFVRYCFSEMAPVCAVVGGILAQEIVKALSQRDPPHNNFFFFDGMKGSGIV 344
> cel:C08B6.9 aos-1; Activator Of Sumo (yeast AOS homolog) family
member (aos-1); K10684 ubiquitin-like 1-activating enzyme
E1 A [EC:6.3.2.19]
Length=343
Score = 38.9 bits (89), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 230 GSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGL 273
G F TAA VG V QEAIK +++ P+ N F++ G + G
Sbjct 293 GPNFGPTAACVGGVIGQEAIKSISEGKNPLRNLFIYTGFESTGF 336
> dre:415148 sae1, sae2a, uble1a, wu:fa28b04, zgc:86633; SUMO1
activating enzyme subunit 1; K10684 ubiquitin-like 1-activating
enzyme E1 A [EC:6.3.2.19]
Length=348
Score = 38.1 bits (87), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 225 LVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLT 274
VSY SE A+VG V QE +K L+++ P N F ++G++ G+
Sbjct 293 FVSYCFSEMSPVCAVVGGVLGQEIVKALSQRDAPHRNFFFFDGLKGSGVV 342
> tgo:TGME49_011390 hypothetical protein ; K10684 ubiquitin-like
1-activating enzyme E1 A [EC:6.3.2.19]
Length=405
Score = 36.6 bits (83), Expect = 0.094, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 230 GSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWN 266
G ++ TAA+VG + AQE K + K+ EPI N ++N
Sbjct 344 GCQYTVTAAVVGGILAQELRKYITKEQEPIPNCLVFN 380
> xla:443558 sae1, Aos1p, MGC78909, Sua1p, aos; SUMO1 activating
enzyme subunit 1; K10684 ubiquitin-like 1-activating enzyme
E1 A [EC:6.3.2.19]
Length=344
Score = 36.6 bits (83), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 223 QQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQRRGLT 274
+ SY SE A+VG V QE +K L+ + P NN F ++G G+
Sbjct 287 KDFASYCFSEMAPVCAVVGGVLGQEIVKALSLRDAPHNNFFFFDGKTSNGIV 338
> dre:567370 ubiquitin-activating enzyme E1-like; K10699 ubiquitin-activating
enzyme E1-like protein 2 [EC:6.3.2.19]
Length=1060
Score = 36.2 bits (82), Expect = 0.15, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 40/76 (52%), Gaps = 2/76 (2%)
Query 195 KDSAAVLLELEEMEKAVGCRLFAAAKVVQQLVS-YRGSEFPATAALVGAVAAQEAIKLLN 253
+DS +L EE+ K + ++ +V+ + RG FP AA VG +A+QE +K L
Sbjct 350 QDSELLLKYTEEISKTLKNKVCINPDLVRCVSRCARGCLFP-LAATVGGIASQEVLKALT 408
Query 254 KKFEPINNTFLWNGVQ 269
KF P+ F + ++
Sbjct 409 GKFSPLQQWFYLDALE 424
> tgo:TGME49_114890 ubiquitin-activating enzyme E1, putative
Length=2759
Score = 34.3 bits (77), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 28/44 (63%), Gaps = 2/44 (4%)
Query 228 YRGSE--FPATAALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQ 269
YR +E A+++GA+AAQEAIK L+ +F P + F ++ +
Sbjct 1179 YRSAEGHLAPIASIMGALAAQEAIKALSGRFTPFHQFFYFDALD 1222
> tpv:TP02_0331 ubiquitin activating enzyme, putatuve
Length=1126
Score = 34.3 bits (77), Expect = 0.48, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 220 KVVQQLVSYRGSEFPATAALVGAVAAQEAIKLLNKKFEP 258
KVV + + PA +L+GA+AAQE IK + F+P
Sbjct 418 KVVSNYNCLKNFKIPAMNSLIGALAAQECIKSITHAFKP 456
> cpv:cgd8_160 ubiquitin activating enzyme E1 ; K04532 amyloid
beta precursor protein binding protein 1
Length=509
Score = 32.7 bits (73), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 36/63 (57%), Gaps = 2/63 (3%)
Query 12 ADTKSYLELQKIYNEKAERDERRL-RELLGQLDVHPSA-KLNVSQEEIKCFCKNAVFFKV 69
+T SYL+LQKI++E+ D R+ G++++ S LN+S E ++ C++ +
Sbjct 358 CETVSYLQLQKIFHEQYTLDVSRISNSNTGKMNISNSDFDLNISDELVQFVCRHLYCLRF 417
Query 70 TQY 72
++
Sbjct 418 IEF 420
> mmu:74153 Uba7, 1300004C08Rik, Ube1l; ubiquitin-like modifier
activating enzyme 7; K10698 ubiquitin-activating enzyme E1-like
[EC:6.3.2.19]
Length=977
Score = 32.3 bits (72), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 12/33 (36%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 237 AALVGAVAAQEAIKLLNKKFEPINNTFLWNGVQ 269
AA++G VAAQE +K +++KF P++ ++ ++
Sbjct 348 AAIMGGVAAQEVLKAISRKFMPLDQWLYFDALE 380
> dre:567793 transmembrane channel-like 3-like
Length=1125
Score = 31.6 bits (70), Expect = 3.0, Method: Composition-based stats.
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 166 GEAAAAATGGAAAARSVEEAIKQQQELVEKDSAAVLLEL 204
G AA + GAA S+ EAI ++QE + S AVL+ L
Sbjct 281 GNLEAAESKGAAIVNSIREAIVEEQEKKKDTSLAVLISL 319
> hsa:7318 UBA7, D8, MGC12713, UBA1B, UBE1L, UBE2; ubiquitin-like
modifier activating enzyme 7; K10698 ubiquitin-activating
enzyme E1-like [EC:6.3.2.19]
Length=1012
Score = 31.6 bits (70), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 237 AALVGAVAAQEAIKLLNKKFEPINNTFLWNGV 268
A++GAVAAQE +K +++KF P++ ++ +
Sbjct 361 VAMLGAVAAQEVLKAISRKFMPLDQWLYFDAL 392
> xla:447263 MGC86332 protein; K01395 meprin A, alpha [EC:3.4.24.18]
Length=705
Score = 30.8 bits (68), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 39/145 (26%), Positives = 59/145 (40%), Gaps = 12/145 (8%)
Query 42 LDVHPSAKLNVSQEEIKCFCKNAVFFKVTQYPKITGAFLGSTSVELLAAAAREMQQAVPE 101
LD S ++ SQ + F KN + + AFL T V + AAA RE Q+ V E
Sbjct 566 LDFGWSTFISHSQLHRRSFLKNDNLILFVDFEDL--AFLSKTEVPIKAAAPRE-QEVVLE 622
Query 102 ELKLSPNADPDLFADPNFSVYEENGFLHLPWYAAALACRAAAAKLGRFPGSHPKPSAAAA 161
K S + DPN + NG + + A +CR ++++ + G +
Sbjct 623 RSKRSAQHFGETLCDPNPCL---NGGVCVQEKGKA-SCRCSSSQAFFYAGEQ-----CES 673
Query 162 TAATGEAAAAATGGAAAARSVEEAI 186
G GG A A ++ AI
Sbjct 674 MKIHGNVLGMLIGGGAGAIALTMAI 698
> pfa:PF10_0057 regulator of nonsense transcripts, putative; K14326
regulator of nonsense transcripts 1 [EC:3.6.4.-]
Length=1554
Score = 30.8 bits (68), Expect = 5.3, Method: Composition-based stats.
Identities = 17/60 (28%), Positives = 32/60 (53%), Gaps = 4/60 (6%)
Query 6 LTADFAADTKSYLELQKIYNEKAERDERRLRELLGQLDVHPSAKLNVSQEEIKCFCKNAV 65
L D A + LEL++ E +++DERRL++L+ + ++ + + I C C A+
Sbjct 884 LKTDIAEELNKLLELKEEVGELSQKDERRLKKLI----LFAEHEILIEADVICCTCVGAM 939
> tgo:TGME49_120680 hypothetical protein
Length=1438
Score = 30.4 bits (67), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 40/157 (25%), Positives = 60/157 (38%), Gaps = 34/157 (21%)
Query 93 REMQQAVPEELKLSPNADP------------DLFADPNFSVYEENGFLHLPW---YAAAL 137
R+ QA+P+ L L+P+A P + +P VY +H P Y +
Sbjct 572 RQALQAMPQPLNLAPHATPFMSRAGGLYDQREASVEPGRDVYP----VHYPTPYAYGPGI 627
Query 138 ACRAAAAKLGRFPGSHPKPSAAAATAATGEAAAAATGGAAAARSVEEAIKQQ---QELVE 194
A A G P H PS A G +A A + RS E Q Q +
Sbjct 628 PADAGAPSAGPGPYPHQFPSGGAGYVVNGRVPDSADHEAHSPRSPESYWGPQAGSQGAED 687
Query 195 KDSAAVLLELEEMEKAVGCRLFAAAKV-VQQLVSYRG 230
KD + VGC L +++ ++++ SY G
Sbjct 688 KDC-----------QVVGCMLPNGSEMAMRRMESYVG 713
> xla:379877 uba1-b, MGC52522, a1s9, a1s9t, a1st, amcx1, gxp1,
poc20, smax2, uba1a, ube1, ube1x; ubiquitin-like modifier activating
enzyme 1 (EC:6.3.2.19); K03178 ubiquitin-activating
enzyme E1 [EC:6.3.2.19]
Length=1059
Score = 30.0 bits (66), Expect = 8.7, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 40/87 (45%), Gaps = 7/87 (8%)
Query 180 RSVEEAIKQQQELVEKDSAAVLLELEEMEKAVGCRLFAAAK-------VVQQLVSYRGSE 232
+ + E K+ L + + A LE+ + +A+ ++K +++QL
Sbjct 333 QGLHEFQKKHGHLPKAHNEADALEVLALTQAINENASGSSKQEEIKESLIKQLAYQATGN 392
Query 233 FPATAALVGAVAAQEAIKLLNKKFEPI 259
A +G +AAQEA+K + KF PI
Sbjct 393 LAPVNAFIGGLAAQEAMKACSGKFMPI 419
Lambda K H
0.316 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10592847112
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40