bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2540_orf1
Length=162
Score E
Sequences producing significant alignments: (Bits) Value
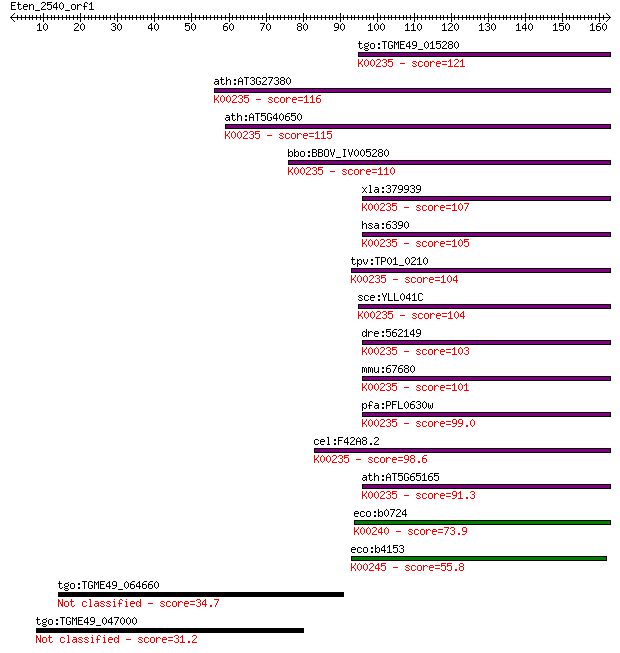
tgo:TGME49_015280 iron-sulfur subunit of succinate dehydrogena... 121 1e-27
ath:AT3G27380 SDH2-1; SDH2-1; electron carrier/ succinate dehy... 116 3e-26
ath:AT5G40650 SDH2-2; SDH2-2; electron carrier/ succinate dehy... 115 5e-26
bbo:BBOV_IV005280 23.m06368; succinate dehydrogenase iron-sulf... 110 2e-24
xla:379939 sdhb, Ip, MGC130803, MGC53699; succinate dehydrogen... 107 2e-23
hsa:6390 SDHB, FLJ92337, IP, PGL4, SDH, SDH1, SDH2, SDHIP; suc... 105 5e-23
tpv:TP01_0210 succinate dehydrogenase iron-sulfur subunit; K00... 104 1e-22
sce:YLL041C SDH2, ACN17; Iron-sulfur protein subunit of succin... 104 1e-22
dre:562149 sdhb, Ip, MGC165456, fj46d06, wu:fj46d06; succinate... 103 3e-22
mmu:67680 Sdhb, 0710008N11Rik; succinate dehydrogenase complex... 101 9e-22
pfa:PFL0630w iron-sulfur subunit of succinate dehydrogenase (E... 99.0 6e-21
cel:F42A8.2 sdhb-1; Succinate DeHydrogenase complex subunit B ... 98.6 7e-21
ath:AT5G65165 SDH2-3; SDH2-3; electron carrier/ succinate dehy... 91.3 1e-18
eco:b0724 sdhB, ECK0713, JW0714; succinate dehydrogenase, FeS ... 73.9 2e-13
eco:b4153 frdB, ECK4149, JW4114; fumarate reductase (anaerobic... 55.8 6e-08
tgo:TGME49_064660 SRS domain-containing, N-acetylglucosamine-p... 34.7 0.14
tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72) 31.2 1.7
> tgo:TGME49_015280 iron-sulfur subunit of succinate dehydrogenase,
putative (EC:1.3.5.1); K00235 succinate dehydrogenase
(ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=342
Score = 121 bits (303), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 52/68 (76%), Positives = 60/68 (88%), Gaps = 0/68 (0%)
Query 95 TFRIHRYDPEKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
TF I+RY+PE RP+MQ++ LD STCGPM+LDALIAIKDRQDP+L FRRSCREGICGSC
Sbjct 44 TFAIYRYNPETDKRPYMQKFELDVSTCGPMILDALIAIKDRQDPSLVFRRSCREGICGSC 103
Query 155 AMNIDGIN 162
AMN+DG N
Sbjct 104 AMNVDGKN 111
> ath:AT3G27380 SDH2-1; SDH2-1; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=279
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 55/107 (51%), Positives = 74/107 (69%), Gaps = 2/107 (1%)
Query 56 PAGVSAACRLLFPRASPVLSGLVSQQRASFSSNAGGTNLTFRIHRYDPEKGGRPHMQEYT 115
P+ ++ A RL+ R + +G ++ +AS G TF+I+R++P+ G+P +Q Y
Sbjct 14 PSKLATAARLIPARWTS--TGAEAETKASSGGGRGSNLKTFQIYRWNPDNPGKPELQNYQ 71
Query 116 LDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSCAMNIDGIN 162
+D CGPMVLDALI IK+ DP+LTFRRSCREGICGSCAMNIDG N
Sbjct 72 IDLKDCGPMVLDALIKIKNEMDPSLTFRRSCREGICGSCAMNIDGCN 118
> ath:AT5G40650 SDH2-2; SDH2-2; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=280
Score = 115 bits (289), Expect = 5e-26, Method: Compositional matrix adjust.
Identities = 58/105 (55%), Positives = 77/105 (73%), Gaps = 5/105 (4%)
Query 59 VSAACRLLFPRASPVLSGLVSQQRASFSSNAGGTNL-TFRIHRYDPEKGGRPHMQEYTLD 117
+S A RL+ R + +G +Q +AS + GG +L TF+I+R++P+ G+P +Q+Y +D
Sbjct 17 LSTAARLIPARWTS--TGSEAQSKAS--TGGGGASLKTFQIYRWNPDNPGKPELQDYKID 72
Query 118 TSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSCAMNIDGIN 162
CGPMVLDALI IK+ DP+LTFRRSCREGICGSCAMNIDG N
Sbjct 73 LKDCGPMVLDALIKIKNEMDPSLTFRRSCREGICGSCAMNIDGCN 117
> bbo:BBOV_IV005280 23.m06368; succinate dehydrogenase iron-sulfur
subunit (EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=281
Score = 110 bits (276), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 51/87 (58%), Positives = 63/87 (72%), Gaps = 4/87 (4%)
Query 76 GLVSQQRASFSSNAGGTNLTFRIHRYDPEKGGRPHMQEYTLDTSTCGPMVLDALIAIKDR 135
GL+ + S+N +TF + RY PE G RP MQ Y +DT+ CGPM+LDALI IK+
Sbjct 7 GLIRRCARRLSANG----VTFSVFRYSPESGKRPRMQSYKVDTAACGPMILDALIKIKNE 62
Query 136 QDPTLTFRRSCREGICGSCAMNIDGIN 162
QD TL+FRRSCREGICGSCAMN++G N
Sbjct 63 QDSTLSFRRSCREGICGSCAMNVNGEN 89
> xla:379939 sdhb, Ip, MGC130803, MGC53699; succinate dehydrogenase
complex, subunit B, iron sulfur (Ip) (EC:1.3.5.1); K00235
succinate dehydrogenase (ubiquinone) iron-sulfur protein
[EC:1.3.5.1]
Length=282
Score = 107 bits (266), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 49/68 (72%), Positives = 57/68 (83%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R+DP+K G +P MQ Y +D +TCGPMVLDALI IK+ DPTLTFRRSCREGICGSC
Sbjct 44 FAIYRWDPDKPGDKPRMQTYEVDLNTCGPMVLDALIKIKNEVDPTLTFRRSCREGICGSC 103
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 104 AMNINGGN 111
> hsa:6390 SDHB, FLJ92337, IP, PGL4, SDH, SDH1, SDH2, SDHIP; succinate
dehydrogenase complex, subunit B, iron sulfur (Ip)
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=280
Score = 105 bits (263), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 48/68 (70%), Positives = 56/68 (82%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEKGG-RPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R+DP+K G +PHMQ Y +D + CGPMVLDALI IK+ D TLTFRRSCREGICGSC
Sbjct 42 FAIYRWDPDKAGDKPHMQTYEVDLNKCGPMVLDALIKIKNEVDSTLTFRRSCREGICGSC 101
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 102 AMNINGGN 109
> tpv:TP01_0210 succinate dehydrogenase iron-sulfur subunit; K00235
succinate dehydrogenase (ubiquinone) iron-sulfur protein
[EC:1.3.5.1]
Length=263
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 46/70 (65%), Positives = 54/70 (77%), Gaps = 0/70 (0%)
Query 93 NLTFRIHRYDPEKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICG 152
N+ F + RY P +P MQ YT+DT+ CGPM+LDALI IK+ D TLTFRRSCREGICG
Sbjct 20 NVNFSVFRYTPNSKEKPRMQTYTVDTNECGPMILDALIKIKNEMDSTLTFRRSCREGICG 79
Query 153 SCAMNIDGIN 162
SCAMNI+G N
Sbjct 80 SCAMNINGEN 89
> sce:YLL041C SDH2, ACN17; Iron-sulfur protein subunit of succinate
dehydrogenase (Sdh1p, Sdh2p, Sdh3p, Sdh4p), which couples
the oxidation of succinate to the transfer of electrons
to ubiquinone as part of the TCA cycle and the mitochondrial
respiratory chain (EC:1.3.5.1); K00235 succinate dehydrogenase
(ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=266
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 46/69 (66%), Positives = 56/69 (81%), Gaps = 1/69 (1%)
Query 95 TFRIHRYDP-EKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGS 153
TF+++R++P E +PH+Q Y +D + CGPMVLDAL+ IKD QD TLTFRRSCREGICGS
Sbjct 35 TFKVYRWNPDEPSAKPHLQSYQVDLNDCGPMVLDALLKIKDEQDSTLTFRRSCREGICGS 94
Query 154 CAMNIDGIN 162
CAMNI G N
Sbjct 95 CAMNIGGRN 103
> dre:562149 sdhb, Ip, MGC165456, fj46d06, wu:fj46d06; succinate
dehydrogenase complex, subunit B, iron sulfur (Ip) (EC:1.3.5.1);
K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=280
Score = 103 bits (257), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 47/68 (69%), Positives = 56/68 (82%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F+I+R+DP+ G +P MQ Y +D +TCGPMVLDALI IK+ D TLTFRRSCREGICGSC
Sbjct 41 FQIYRWDPDTVGDKPRMQTYEIDLNTCGPMVLDALIKIKNEMDSTLTFRRSCREGICGSC 100
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 101 AMNINGGN 108
> mmu:67680 Sdhb, 0710008N11Rik; succinate dehydrogenase complex,
subunit B, iron sulfur (Ip) (EC:1.3.5.1); K00235 succinate
dehydrogenase (ubiquinone) iron-sulfur protein [EC:1.3.5.1]
Length=282
Score = 101 bits (252), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 47/68 (69%), Positives = 55/68 (80%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F I+R+DP+K G +P MQ Y +D + CGPMVLDALI IK+ D TLTFRRSCREGICGSC
Sbjct 44 FAIYRWDPDKTGDKPRMQTYEVDLNKCGPMVLDALIKIKNEVDSTLTFRRSCREGICGSC 103
Query 155 AMNIDGIN 162
AMNI+G N
Sbjct 104 AMNINGGN 111
> pfa:PFL0630w iron-sulfur subunit of succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone) iron-sulfur
protein [EC:1.3.5.1]
Length=321
Score = 99.0 bits (245), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 43/67 (64%), Positives = 49/67 (73%), Gaps = 0/67 (0%)
Query 96 FRIHRYDPEKGGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSCA 155
F I RY+P RP M+ + +D CGPMVLD LI IKD D TL+FRRSCREGICGSCA
Sbjct 70 FSIFRYNPTNKKRPQMETFEVDIDNCGPMVLDVLIKIKDEIDSTLSFRRSCREGICGSCA 129
Query 156 MNIDGIN 162
MNI+G N
Sbjct 130 MNINGKN 136
> cel:F42A8.2 sdhb-1; Succinate DeHydrogenase complex subunit
B family member (sdhb-1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=298
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 46/81 (56%), Positives = 58/81 (71%), Gaps = 1/81 (1%)
Query 83 ASFSSNAGGTNLTFRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLT 141
A+ + G TF I+R++PE G +P +Q++ +D CG M+LDALI IK+ DPTLT
Sbjct 43 AAKTKKTGNRIKTFEIYRFNPEAPGAKPTVQKFDVDLDQCGTMILDALIKIKNEVDPTLT 102
Query 142 FRRSCREGICGSCAMNIDGIN 162
FRRSCREGICGSCAMNI G N
Sbjct 103 FRRSCREGICGSCAMNIGGQN 123
> ath:AT5G65165 SDH2-3; SDH2-3; electron carrier/ succinate dehydrogenase
(EC:1.3.5.1); K00235 succinate dehydrogenase (ubiquinone)
iron-sulfur protein [EC:1.3.5.1]
Length=309
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/68 (57%), Positives = 53/68 (77%), Gaps = 1/68 (1%)
Query 96 FRIHRYDPEK-GGRPHMQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGICGSC 154
F+I+R++P+K +P +Q + +D S+CGPMVLD L IK D +L++RRSCREGICGSC
Sbjct 69 FKIYRWNPDKPNSKPFLQSFFVDLSSCGPMVLDVLQKIKAEDDASLSYRRSCREGICGSC 128
Query 155 AMNIDGIN 162
+MNIDG N
Sbjct 129 SMNIDGTN 136
> eco:b0724 sdhB, ECK0713, JW0714; succinate dehydrogenase, FeS
subunit (EC:1.3.99.1); K00240 succinate dehydrogenase iron-sulfur
protein [EC:1.3.99.1]
Length=238
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 36/70 (51%), Positives = 51/70 (72%), Gaps = 2/70 (2%)
Query 94 LTFRIHRYDPEKGGRPHMQEYTLDTSTC-GPMVLDALIAIKDRQDPTLTFRRSCREGICG 152
L F I+RY+P+ P MQ+YTL+ M+LDALI +K++ DP+L+FRRSCREG+CG
Sbjct 3 LEFSIYRYNPDVDDAPRMQDYTLEADEGRDMMLLDALIQLKEK-DPSLSFRRSCREGVCG 61
Query 153 SCAMNIDGIN 162
S +N++G N
Sbjct 62 SDGLNMNGKN 71
> eco:b4153 frdB, ECK4149, JW4114; fumarate reductase (anaerobic),
Fe-S subunit (EC:1.3.99.1); K00245 fumarate reductase iron-sulfur
protein [EC:1.3.99.1]
Length=244
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Query 93 NLTFRIHRYDPEKGGRPH--MQEYTLDTSTCGPMVLDALIAIKDRQDPTLTFRRSCREGI 150
NL + RY+PE PH E D +T +LDAL IKD P L++R SCR I
Sbjct 6 NLKIEVVRYNPEVDTAPHSAFYEVPYDATTS---LLDALGYIKDNLAPDLSYRWSCRMAI 62
Query 151 CGSCAMNIDGI 161
CGSC M ++ +
Sbjct 63 CGSCGMMVNNV 73
> tgo:TGME49_064660 SRS domain-containing, N-acetylglucosamine-phosphate
mutase, putative
Length=2344
Score = 34.7 bits (78), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 22/80 (27%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Query 14 CFRQMHAGDSGAVCTLSEAQPSEW--PVNSLIFMEMRRLTSSL-SPAGVSAACRLLFPRA 70
CF + GD C + P W PV + +++ LT ++ SP V+ C L FP+
Sbjct 1866 CFACVDRGDITNSCGVLVTVPPFWAPPVQETVCNDVKTLTMTVHSPGSVTLRCGLEFPKV 1925
Query 71 SPVLSGLVSQQRASFSSNAG 90
P + SQ + + + G
Sbjct 1926 DPPIDDPASQGQVYYGKSCG 1945
> tgo:TGME49_047000 TPR domain-containing protein (EC:3.4.21.72)
Length=888
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 6/72 (8%)
Query 8 DRKVLSCFRQMHAGDSGAVCTLSEAQPSEWPVNSLIFMEMRRLTSSLSPAGVSAACRLLF 67
DR++ CF+ HA + + S + P+ S R SSL P+G + +
Sbjct 244 DRRIRGCFKYDHAESRPSSSSTSSSFRVSSPLQS------RSDPSSLLPSGSAHVTKSEC 297
Query 68 PRASPVLSGLVS 79
PRA PVL+ S
Sbjct 298 PRAPPVLAAPTS 309
Lambda K H
0.320 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3767900632
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40