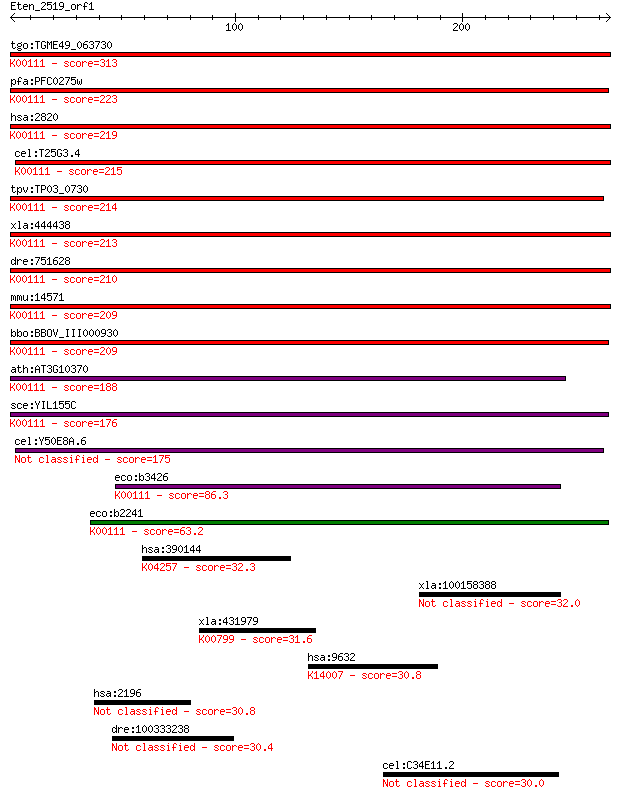
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2519_orf1
Length=264
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative... 313 5e-85
pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,... 223 5e-58
hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydro... 219 1e-56
cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate ... 215 1e-55
tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21)... 214 3e-55
xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase ... 213 7e-55
dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe0... 210 4e-54
mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH, ... 209 7e-54
bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phospha... 209 1e-53
ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosp... 188 1e-47
sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosph... 176 6e-44
cel:Y50E8A.6 hypothetical protein 175 2e-43
eco:b3426 glpD, ECK3412, glyD, JW3389; sn-glycerol-3-phosphate... 86.3 1e-16
eco:b2241 glpA, ECK2233, JW2235; sn-glycerol-3-phosphate dehyd... 63.2 1e-09
hsa:390144 OR5D16, OR11-154; olfactory receptor, family 5, sub... 32.3 1.7
xla:100158388 cyp2j2; cytochrome P450, family 2, subfamily J, ... 32.0 2.4
xla:431979 hypothetical protein MGC82327; K00799 glutathione S... 31.6 3.3
hsa:9632 SEC24C, KIAA0079; SEC24 family, member C (S. cerevisi... 30.8 4.7
hsa:2196 FAT2, CDHF8, CDHR9, HFAT2, MEGF1; FAT tumor suppresso... 30.8 5.3
dre:100333238 protocadherin 2A2-like 30.4 7.7
cel:C34E11.2 hypothetical protein 30.0 9.3
> tgo:TGME49_063730 glycerol-3-phosphate dehydrogenase, putative
(EC:1.1.3.21); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=653
Score = 313 bits (801), Expect = 5e-85, Method: Compositional matrix adjust.
Identities = 146/278 (52%), Positives = 194/278 (69%), Gaps = 14/278 (5%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQD 60
S LYFDGQMNDSRM L LAL+ ++ G+VEGM+ A NH+ + +K+ NG+ +VQD
Sbjct 222 SLLYFDGQMNDSRMNLQLALTASLDGYVEGMKGACVGNHLNVVDILKDGNGKAAGARVQD 281
Query 61 RETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLL 120
RET EEF IH K VVNC GP +D+VR M LV PAAG HIVLPHWYT +P+GLL
Sbjct 282 RETGEEFDIHSKTVVNCAGPFSDAVRTMADPTQPKLVEPAAGIHIVLPHWYTSASPYGLL 341
Query 121 LPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMR 180
LP+T+DGRVLF+LPW+G T+AGTTDAP + P+ EV ++ EL++YL+MD +++R
Sbjct 342 LPKTTDGRVLFMLPWQGATIAGTTDAPCELSDAPQATEEEVAWVTRELASYLKMDEKRLR 401
Query 181 GDIQSVWAGHRPLISQTSEAKD--------------TAGVVRSHTVLVDHESGLISLMGG 226
D++SVW G RPLIS + + TA VVRSH ++VD ++GL+S++GG
Sbjct 402 DDVRSVWKGIRPLISHIPQPNEEAVTAAATKDAQVSTANVVRSHYIVVDEKTGLVSVLGG 461
Query 227 KWTTYRRMAQDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
KWTTYRRMA++ +D LL H+DKVA + CRTK + +Q
Sbjct 462 KWTTYRRMAEETLDALLAAHRDKVATTRSCRTKSLLIQ 499
> pfa:PFC0275w FAD-dependent glycerol-3-phosphate dehydrogenase,
putative; K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=653
Score = 223 bits (568), Expect = 5e-58, Method: Compositional matrix adjust.
Identities = 110/300 (36%), Positives = 168/300 (56%), Gaps = 39/300 (13%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQD 60
S +YFDGQ +DSRM L+L L+ + +V G A NH+ K FIK+EN +++ V+ D
Sbjct 208 SLIYFDGQHDDSRMNLNLILTSAIDNYVPGQIGATVCNHMEVKNFIKDENNKLVGVRAID 267
Query 61 RETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLL 120
+ D+E I KV++N TGP D +RK+ E+ ++ + G H +LP WY+ K G++
Sbjct 268 KINDKEIEIFSKVIINATGPYGDIIRKLADENRKPMIQVSVGCHFILPKWYSTKNN-GMI 326
Query 121 LPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMR 180
+P+TSD RVLFLLPWE T+ GTTD +P+ +++FL NELS Y+ + ++++
Sbjct 327 IPKTSDDRVLFLLPWENNTLVGTTDEKRIMQDDPKIQEKDIEFLTNELSKYIHVSAEEIK 386
Query 181 GDIQSVWAGHRPLISQTSEAKD-------------------------------------T 203
DI + W G RPLIS + K T
Sbjct 387 NDITAAWCGFRPLISTNNNKKSKFSSNKKKNNNNNINSNSSSNSNNFDDEQNNNNNNIGT 446
Query 204 AGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDTLLEVHKDKVAASMPCRTKGMKV 263
+ RSH ++ D ++GLIS++GGKWT YR+MAQD +D +L + +++ CRTK +K+
Sbjct 447 HEISRSHEIIED-DNGLISILGGKWTIYRKMAQDTLDYILRKYPNELKTKYNCRTKFLKL 505
> hsa:2820 GPD2, GDH2, GPDM, mGPDH; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 219 bits (557), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 115/269 (42%), Positives = 168/269 (62%), Gaps = 18/269 (6%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKV-- 58
+ +Y+DGQ ND+RM L++AL+ G AA AN++ +K + Q V+V
Sbjct 215 AIVYYDGQHNDARMNLAIALTAARYG-------AATANYMEVVSLLKKTDPQTGKVRVSG 267
Query 59 ---QDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKT 115
+D T +EF + K V+N TGP TDSVRKMD +DAA++ P+AG HIV+P +Y+ ++
Sbjct 268 ARCKDVLTGQEFDVRAKCVINATGPFTDSVRKMDDKDAAAICQPSAGVHIVMPGYYSPES 327
Query 116 PFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMD 175
GLL P TSDGRV+F LPW+ T+AGTTD P + P P +++F++NE+ YL D
Sbjct 328 -MGLLDPATSDGRVIFFLPWQKMTIAGTTDTPTDVTHHPIPSEEDINFILNEVRNYLSCD 386
Query 176 PQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMA 235
+ RGD+ + W+G RPL++ A DT + R+H V + ESGLI++ GGKWTTYR MA
Sbjct 387 VEVRRGDVLAAWSGIRPLVTDPKSA-DTQSISRNHVVDIS-ESGLITIAGGKWTTYRSMA 444
Query 236 QDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
+D ++ ++ H K P RT G+ +Q
Sbjct 445 EDTINAAVKTHNLKAG---PSRTVGLFLQ 470
> cel:T25G3.4 hypothetical protein; K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=722
Score = 215 bits (547), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 115/263 (43%), Positives = 165/263 (62%), Gaps = 13/263 (4%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
+Y+DGQ ND+RM L++ L+ G AA ANHV ++ K+E G++I V+D
Sbjct 222 IYYDGQHNDARMNLAIILTAIRHG-------AACANHVRVEKLNKDETGKVIGAHVRDMV 274
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKM-DHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLL 121
T E+ I K V+N TGP TDS+R M D E A + P++G HI LP +Y+ + GLL
Sbjct 275 TGGEWDIKAKAVINATGPFTDSIRLMGDPETARPICAPSSGVHITLPGYYS-PSNTGLLD 333
Query 122 PQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRG 181
P TSDGRV+F LPWE T+AGTTDAP++ P+P +++F++ E+ YL D RG
Sbjct 334 PDTSDGRVIFFLPWERMTIAGTTDAPSDVTLSPQPTDHDIEFILQEIRGYLSKDVSVRRG 393
Query 182 DIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDT 241
D+ S W+G RPL+ ++ KDT + R+H + V +SGLI++ GGKWTTYR MA++ VD
Sbjct 394 DVMSAWSGLRPLVRDPNK-KDTKSLARNHIIEVG-KSGLITIAGGKWTTYRHMAEETVDR 451
Query 242 LLEVHKDKVAASMPCRTKGMKVQ 264
++EVH K C T G+ ++
Sbjct 452 VVEVHGLKTENG--CVTPGLLLE 472
> tpv:TP03_0730 glycerol-3-phosphate dehydrogenase (EC:1.1.3.21);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=615
Score = 214 bits (544), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 103/262 (39%), Positives = 162/262 (61%), Gaps = 4/262 (1%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN-ENGQIIAVKVQ 59
+ +Y+DGQ NDSR L +AL+ TV +V G + + N+V + +++ + V+
Sbjct 210 AVVYYDGQHNDSRTNLMMALTSTVNNYVPGQKGSTVCNYVKVNKILRDPATNSVNGAVVE 269
Query 60 DRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGL 119
D T EEFTI+ KVVVNCTGP ++++ + ++ + GTH+ LP Y TP+GL
Sbjct 270 DVLTGEEFTINSKVVVNCTGPFAGETKRLNDPGSDLSILHSRGTHLTLPRKYC-PTPYGL 328
Query 120 LLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQM 179
++P+T+DGRVLF+LPW T+ GTTD + A P+P +V+F+ + S Y+ +Q+
Sbjct 329 IIPKTTDGRVLFVLPWLNETIIGTTDNKDDLACNPKPTKEDVEFITKDASIYMNCPDEQL 388
Query 180 RGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAV 239
+ +I+S+W+G RPLI E T + R H + VD ++G++++ GGKWT R MAQD V
Sbjct 389 KSEIKSLWSGLRPLIKGVDEVNQTGKLSRGHIIEVD-KTGMVNVYGGKWTICRLMAQDCV 447
Query 240 DTLLEVHKDKVAASMPCRTKGM 261
D LL+ +KD + S CRT+ +
Sbjct 448 DELLKFYKD-LKQSYKCRTRNL 468
> xla:444438 gpd2, MGC83596; glycerol-3-phosphate dehydrogenase
2 (mitochondrial) (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=725
Score = 213 bits (541), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 111/269 (41%), Positives = 165/269 (61%), Gaps = 18/269 (6%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN---ENGQ--IIA 55
+ +Y+DGQ ND+RM L++AL+ G AA AN+ ++ E+G+ +
Sbjct 213 AIVYYDGQHNDARMNLAIALTAARYG-------AATANYTEVVRLLRKIDPESGKELVCG 265
Query 56 VKVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKT 115
+ +D T +EF + K V+N TGP TDSVRKMD ++ ++ P+AG HIV+P +Y+
Sbjct 266 ARCRDILTGQEFDVRAKCVINATGPFTDSVRKMDDQEVKNICQPSAGVHIVMPGYYSPDN 325
Query 116 PFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMD 175
GLL P TSDGRV+F LPWE T+AGTTD P P P +++F++ E+ YL D
Sbjct 326 -MGLLDPATSDGRVIFFLPWEKMTIAGTTDTPTEVTHHPIPTDEDINFILTEVRNYLSSD 384
Query 176 PQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMA 235
+ RGD+ + W+G RPL++ + +KDT + R+H V V ESGL+++ GGKWTTYR MA
Sbjct 385 VEVRRGDVLAAWSGIRPLVTNPN-SKDTQSISRNHVVDVS-ESGLVTIAGGKWTTYRSMA 442
Query 236 QDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
+D +D ++ H K PCRT G+ ++
Sbjct 443 EDTLDAAVKAHNLKAG---PCRTVGLFLE 468
> dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe02f02,
zgc:153729; glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
(EC:1.1.5.3); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=536
Score = 210 bits (535), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 112/269 (41%), Positives = 167/269 (62%), Gaps = 18/269 (6%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN---ENGQ--IIA 55
+ +Y+DGQ ND+RM L++AL+ G AA AN+ +K + GQ +
Sbjct 214 AIVYYDGQHNDARMNLAIALTAARHG-------AAIANYTEVVHLLKKPDPKTGQEKVCG 266
Query 56 VKVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKT 115
+ +D T +EF I K V+N TGP TDS+RKMD++ A++ P+AG HIV+P +Y+
Sbjct 267 ARCRDVVTGQEFDIRAKCVINATGPFTDSLRKMDNQKTANICQPSAGVHIVIPGYYSPDN 326
Query 116 PFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMD 175
GLL P TSDGRV+F LPWE T+AGTTD P A P P +++F+++E+ YL D
Sbjct 327 -MGLLDPATSDGRVIFFLPWEKMTIAGTTDTPTEVTAHPIPMEDDINFILSEVRNYLSPD 385
Query 176 PQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMA 235
+ RGD+ + W+G RPL++ S +KDT + R+H V V +SGLI++ GGKWTTYR MA
Sbjct 386 VEVRRGDVLAAWSGIRPLVTDPS-SKDTQSICRNHIVNVS-DSGLITIAGGKWTTYRSMA 443
Query 236 QDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
++ +D ++ H ++ P RT G+ ++
Sbjct 444 EETLDAAIKAHN---LSAGPSRTVGLTLE 469
> mmu:14571 Gpd2, AA408484, AI448216, AU021455, AW494132, GPDH,
Gdm1, Gpd-m, Gpdh-m, TISP38; glycerol phosphate dehydrogenase
2, mitochondrial (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=727
Score = 209 bits (533), Expect = 7e-54, Method: Compositional matrix adjust.
Identities = 111/269 (41%), Positives = 163/269 (60%), Gaps = 18/269 (6%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN---ENGQ--IIA 55
+ +Y+DGQ ND+RM L++AL+ G AA AN++ +K E G+ +
Sbjct 215 AIVYYDGQHNDARMNLAIALTAARYG-------AATANYMEVVSLLKKTDPETGKERVSG 267
Query 56 VKVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKT 115
+ +D T +EF + K V+N +GP TDSVRKMD ++ + P+AG HIV+P +Y+ +
Sbjct 268 ARCKDVLTGQEFDVRAKCVINASGPFTDSVRKMDDKNVVPICQPSAGVHIVMPGYYSPEN 327
Query 116 PFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMD 175
GLL P TSDGRV+F LPWE T+AGTTD P + P P +++F++NE+ YL D
Sbjct 328 -MGLLDPATSDGRVIFFLPWEKMTIAGTTDTPTDVTHHPIPSEEDINFILNEVRNYLSSD 386
Query 176 PQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMA 235
+ RGD+ + W+G RPL++ A DT + R+H V + +SGLI++ GGKWTTYR MA
Sbjct 387 VEVRRGDVLAAWSGIRPLVTDPKSA-DTQSISRNHVVDIS-DSGLITIAGGKWTTYRSMA 444
Query 236 QDAVDTLLEVHKDKVAASMPCRTKGMKVQ 264
+D VD ++ H P RT G+ +Q
Sbjct 445 EDTVDAAVKFHNLNAG---PSRTVGLFLQ 470
> bbo:BBOV_III000930 17.m07109; FAD-dependent glycerol-3-phosphate
dehydrogenase (EC:1.1.99.5); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=619
Score = 209 bits (531), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 104/266 (39%), Positives = 164/266 (61%), Gaps = 6/266 (2%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQD 60
+ +Y+DGQ +DSR L +AL+ T+ +V G A NH E +K+ G++ KV D
Sbjct 210 AVVYYDGQHDDSRTNLLMALTSTIDNYVPGQVGATVVNHTEVVELLKDSEGKVCGAKVVD 269
Query 61 RETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLL 120
+ T + F + K VVNC+GP + VR++ + D + ++ + GTHIVLP Y+ T FG++
Sbjct 270 KLTGDTFDVRAKAVVNCSGPFAEKVRQIGNPDGSENMLHSRGTHIVLPATYS-PTQFGMV 328
Query 121 LPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMR 180
+P+T+DGRVLF LPW G+T+ GTTD + P P S+VDF+ + + Y+ + +R
Sbjct 329 IPKTTDGRVLFTLPWRGQTLVGTTDNKDDLQWNPLPKKSDVDFICKDAAIYMNCSEESLR 388
Query 181 GDIQSVWAGHRPLISQTS---EAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQD 237
DI+SVW+G RPL+ +++ T + R H + VD ++ L+++ GGKWT R MA++
Sbjct 389 RDIKSVWSGLRPLLKGLDGQPDSEKTDSLSRGHVIHVDRQN-LVNVYGGKWTICRLMAEE 447
Query 238 AVDTLLEVHKDKVAASMPCRTKGMKV 263
VD LLE + D V A CRT+ +++
Sbjct 448 CVDRLLEANPD-VKAKTRCRTRNLRL 472
> ath:AT3G10370 SDP6; SDP6 (SUGAR-DEPENDENT 6); glycerol-3-phosphate
dehydrogenase (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=629
Score = 188 bits (478), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 101/245 (41%), Positives = 151/245 (61%), Gaps = 13/245 (5%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNE-NGQIIAVKVQ 59
+ +Y+DGQMNDSR+ + LA + + AA NH I ++ +II +++
Sbjct 223 TVVYYDGQMNDSRLNVGLACTAA-------LAGAAVLNHAEVVSLITDDATKRIIGARIR 275
Query 60 DRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGL 119
+ T +EF + KVVVN GP DS+RKM ED ++ P++G HIVLP +Y+ + GL
Sbjct 276 NNLTGQEFNSYAKVVVNAAGPFCDSIRKMIDEDTKPMICPSSGVHIVLPDYYSPEG-MGL 334
Query 120 LLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQM 179
++P+T DGRV+F+LPW GRTVAGTTD+ + + P P E+ F+++ +S YL + +
Sbjct 335 IVPKTKDGRVVFMLPWLGRTVAGTTDSNTSITSLPEPHEDEIQFILDAISDYLNIKVR-- 392
Query 180 RGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAV 239
R D+ S W+G RPL + AK T + R H V + GL+++ GGKWTTYR MA+DAV
Sbjct 393 RTDVLSAWSGIRPL-AMDPTAKSTESISRDHVVF-EENPGLVTITGGKWTTYRSMAEDAV 450
Query 240 DTLLE 244
D ++
Sbjct 451 DAAIK 455
> sce:YIL155C GUT2; Gut2p (EC:1.1.5.3); K00111 glycerol-3-phosphate
dehydrogenase [EC:1.1.5.3]
Length=649
Score = 176 bits (447), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 110/299 (36%), Positives = 163/299 (54%), Gaps = 52/299 (17%)
Query 1 STLYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKN-ENGQIIAVKVQ 59
S +Y DG NDSR+ +LA++ G A N+V ++ IK+ +G++I + +
Sbjct 213 SLVYHDGSFNDSRLNATLAITAVENG-------ATVLNYVEVQKLIKDPTSGKVIGAEAR 265
Query 60 DRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAA------------------------S 95
D ET+E I+ K VVN TGP +D++ +MD +
Sbjct 266 DVETNELVRINAKCVVNATGPYSDAILQMDRNPSGLPDSPLNDNSKIKSTFNQIAVMDPK 325
Query 96 LVIPAAGTHIVLPHWYTHKTPFGLLLPQTSDGRVLFLLPWEGRTVAGTTDAPANKAAE-P 154
+VIP+ G HIVLP +Y K GLL +TSDGRV+F LPW+G+ +AGTTD P + E P
Sbjct 326 MVIPSIGVHIVLPSFYCPKD-MGLLDVRTSDGRVMFFLPWQGKVLAGTTDIPLKQVPENP 384
Query 155 RPGVSEVDFLVNELSAYLRMDPQQMRGDIQSVWAGHRPLI-------SQTSEAKDTAGVV 207
P +++ ++ EL Y+ + R D+ S WAG RPL+ + + T GVV
Sbjct 385 MPTEADIQDILKELQHYIEFPVK--REDVLSAWAGVRPLVRDPRTIPADGKKGSATQGVV 442
Query 208 RSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDTLLEV---HKDKVAASMPCRTKGMKV 263
RSH L ++GLI++ GGKWTTYR+MA++ VD ++EV H K PC T+ +K+
Sbjct 443 RSH-FLFTSDNGLITIAGGKWTTYRQMAEETVDKVVEVGGFHNLK-----PCHTRDIKL 495
> cel:Y50E8A.6 hypothetical protein
Length=609
Score = 175 bits (443), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 89/259 (34%), Positives = 146/259 (56%), Gaps = 11/259 (4%)
Query 3 LYFDGQMNDSRMCLSLALSPTVPGFVEGMQPAAAANHVAAKEFIKNENGQIIAVKVQDRE 62
LY+DGQ ND+R+ L +AL+ G A NH +K+ G++ V+D
Sbjct 210 LYYDGQGNDARLVLVVALTAIRNG-------AKCVNHTECIGLLKDSYGKVNGALVKDHI 262
Query 63 TDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLLP 122
+ E + IH KVVVN TGP D +R+M E +++ ++G H+ + ++ GL++P
Sbjct 263 SGETYEIHSKVVVNATGPFNDHIREMADETRNKIIVGSSGIHLTVAKYFCPGNA-GLIVP 321
Query 123 QTSDGRVLFLLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRGD 182
++SDGRV+F PWE T+ GTTD P + P E+ +++ E++ + + R D
Sbjct 322 KSSDGRVIFAFPWENVTIVGTTDDPTEPSHSPTVTEKEIQYILKEMNRAFEKEYEIKRED 381
Query 183 IQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLMGGKWTTYRRMAQDAVDTL 242
+ S+W+G R L+ K+ + R H V V +GLI++ GGK TT+R MA++ V+ L
Sbjct 382 VTSIWSGIRGLVQDPKRQKEHNSLARGHLVDVG-PTGLITIAGGKLTTFRHMAEETVNKL 440
Query 243 LEVHKDKVAASMPCRTKGM 261
L++ +K+ + PC T+GM
Sbjct 441 LDI--NKILEAQPCVTRGM 457
> eco:b3426 glpD, ECK3412, glyD, JW3389; sn-glycerol-3-phosphate
dehydrogenase, aerobic, FAD/NAD(P)-binding (EC:1.1.5.3);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=501
Score = 86.3 bits (212), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 64/208 (30%), Positives = 111/208 (53%), Gaps = 26/208 (12%)
Query 47 KNENGQIIAVKVQDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAA----- 101
+ ENG I V+ +D +T ++++ + +VN TGP K +D L P
Sbjct 176 RRENGLWI-VEAEDIDTGKKYSWQARGLVNATGPWV----KQFFDDGMHLPSPYGIRLIK 230
Query 102 GTHIVLPHWYTHKTPFGLLLPQTSDGRVLFLLPW-EGRTVAGTTDAPANKAAEPRP---G 157
G+HIV+P +T K + L Q D R++F++PW + ++ GTTD +P+
Sbjct 231 GSHIVVPRVHTQKQAYIL---QNEDKRIVFVIPWMDEFSIIGTTDVEYK--GDPKAVKIE 285
Query 158 VSEVDFLVNELSAYLRMDPQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHE 217
SE+++L+N + + + Q R DI ++G RPL S++ + R +T+ + E
Sbjct 286 ESEINYLLNVYNTHFK--KQLSRDDIVWTYSGVRPLCDDESDSPQ--AITRDYTLDIHDE 341
Query 218 SG---LISLMGGKWTTYRRMAQDAVDTL 242
+G L+S+ GGK TTYR++A+ A++ L
Sbjct 342 NGKAPLLSVFGGKLTTYRKLAEHALEKL 369
> eco:b2241 glpA, ECK2233, JW2235; sn-glycerol-3-phosphate dehydrogenase
(anaerobic), large subunit, FAD/NAD(P)-binding (EC:1.1.5.3);
K00111 glycerol-3-phosphate dehydrogenase [EC:1.1.5.3]
Length=542
Score = 63.2 bits (152), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 64/247 (25%), Positives = 105/247 (42%), Gaps = 34/247 (13%)
Query 36 AANHVAAKEF------------IKNENGQIIAVKVQDRETDEEFTIHCKVVVNCTGPLTD 83
AAN + AKE + E + V+V++ T E +H VVVN G
Sbjct 155 AANMLDAKEHGAVILTAHEVTGLIREGATVCGVRVRNHLTGETQALHAPVVVNAAGIWGQ 214
Query 84 SVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFGLLLPQTSDGRVLFLLPWEGRTVAGT 143
+ ++ D + PA G+ +++ H P +D L+P + ++ GT
Sbjct 215 HI--AEYADLRIRMFPAKGSLLIMDHRINQHVINRCRKPSDAD----ILVPGDTISLIGT 268
Query 144 TD--APANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRGDIQSVWAGHRPLISQTSEAK 201
T N+ + R EVD L+ E ++ P + I ++G RPL++ S+
Sbjct 269 TSLRIDYNEIDDNRVTAEEVDILLREGE---KLAPVMAKTRILRAYSGVRPLVA--SDDD 323
Query 202 DTAGVVRSHTVLVDHES-----GLISLMGGKWTTYRRMAQDAVDTLLEVHKDKVAASMPC 256
+ V VL+DH G I++ GGK TYR MA+ A D + K+ + PC
Sbjct 324 PSGRNVSRGIVLLDHAERDGLDGFITITGGKLMTYRLMAEWATDAVCR----KLGNTRPC 379
Query 257 RTKGMKV 263
T + +
Sbjct 380 TTADLAL 386
> hsa:390144 OR5D16, OR11-154; olfactory receptor, family 5, subfamily
D, member 16; K04257 olfactory receptor
Length=328
Score = 32.3 bits (72), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 29/65 (44%), Gaps = 4/65 (6%)
Query 59 QDRETDEEFTIHCKVVVNCTGPLTDSVRKMDHEDAASLVIPAAGTHIVLPHWYTHKTPFG 118
R T + ++ VV+ PL S+R D +DA +I HI HWY PF
Sbjct 268 NSRHTVKVASVFYTVVIPLLNPLIYSLRNKDVKDAIRKIINTKYFHIKHRHWY----PFN 323
Query 119 LLLPQ 123
++ Q
Sbjct 324 FVIEQ 328
> xla:100158388 cyp2j2; cytochrome P450, family 2, subfamily J,
polypeptide 2 (EC:1.14.14.1)
Length=433
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/73 (27%), Positives = 33/73 (45%), Gaps = 11/73 (15%)
Query 181 GDIQSVWAGHRPLISQT----------SEAKDTAGV-VRSHTVLVDHESGLISLMGGKWT 229
GDI ++W GH PL+ + S +++ +G V S + +E G++S G W
Sbjct 6 GDIYTLWLGHTPLVVLSGCKSVRNGLISHSEELSGRPVDSFLQALTNERGIVSTNGHTWK 65
Query 230 TYRRMAQDAVDTL 242
RR + L
Sbjct 66 QQRRFGMMTLRNL 78
> xla:431979 hypothetical protein MGC82327; K00799 glutathione
S-transferase [EC:2.5.1.18]
Length=241
Score = 31.6 bits (70), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 33/60 (55%), Gaps = 9/60 (15%)
Query 84 SVRKMDHEDAASLVIPAAGT-HIVL-------PHWYTHKTPFGLLLP-QTSDGRVLFLLP 134
S+R + A LV+ A G H V+ P W+ K+PFGL+ +TS+G+V++ P
Sbjct 27 SMRFCPYAQRARLVLAAKGIKHEVININLKNKPDWFFEKSPFGLVPSLETSNGQVIYESP 86
> hsa:9632 SEC24C, KIAA0079; SEC24 family, member C (S. cerevisiae);
K14007 protein transport protein SEC24
Length=1094
Score = 30.8 bits (68), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 132 LLPWEGRTVAGTTDAPANKAAEPRPGVSEVDFLVNELSAYLRMDPQQMRGDIQSVWA 188
LLP V TT+ PA +A+E R ++ L N L+ +L + +G +QS+++
Sbjct 951 LLPLTKSPVESTTEPPAVRASEERLSNGDIYLLENGLNLFLWVGASVQQGVVQSLFS 1007
> hsa:2196 FAT2, CDHF8, CDHR9, HFAT2, MEGF1; FAT tumor suppressor
homolog 2 (Drosophila)
Length=4349
Score = 30.8 bits (68), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 38 NHVAAKEFIKNENGQIIAVKVQDRETDEEFTIHCKVVVNCTG 79
N +A+++F N NGQI ++ DRE E I KV+ G
Sbjct 2522 NKLASEKFSINPNGQIATLQKLDRENSTERVIAIKVMARDGG 2563
> dre:100333238 protocadherin 2A2-like
Length=798
Score = 30.4 bits (67), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 28/57 (49%), Gaps = 4/57 (7%)
Query 46 IKNENGQIIAVKVQDRETDEEFTIHCKVVVNCTGPLTDSVRK----MDHEDAASLVI 98
I ENG + A+K D E +++F H + + PL+ +V MD D L++
Sbjct 505 INPENGNLYALKTFDYEIEKDFLFHIEARDSGVPPLSSNVTVHIIIMDQNDNTPLIV 561
> cel:C34E11.2 hypothetical protein
Length=677
Score = 30.0 bits (66), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 18/77 (23%), Positives = 31/77 (40%), Gaps = 7/77 (9%)
Query 165 VNELSAYLRMDPQQMRGDIQSVWAGHRPLISQTSEAKDTAGVVRSHTVLVDHESGLISLM 224
+NE+ Y QM ++ HRPL+ RSH++ D++S
Sbjct 314 INEIEKYTESPAVQMLDKPRAGGRAHRPLLQDVI-------FTRSHSITSDYDSNKEHFD 366
Query 225 GGKWTTYRRMAQDAVDT 241
G W++ + +D T
Sbjct 367 SGSWSSTQSRYKDGRST 383
Lambda K H
0.318 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9852431512
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40