bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2493_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
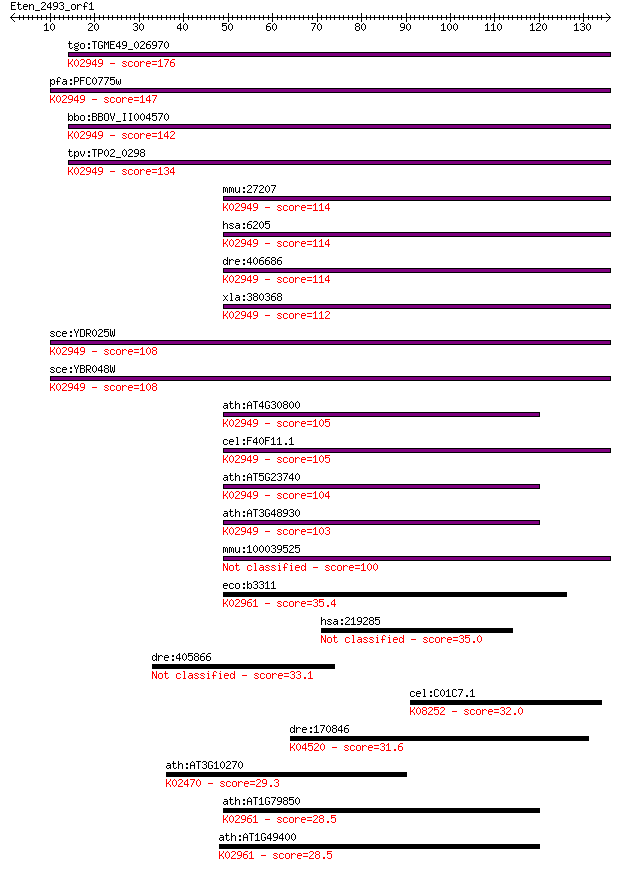
tgo:TGME49_026970 40S ribosomal protein S11, putative ; K02949... 176 1e-44
pfa:PFC0775w 40S ribosomal protein S11, putative; K02949 small... 147 1e-35
bbo:BBOV_II004570 18.m06381; 40S ribosomal protein S11; K02949... 142 3e-34
tpv:TP02_0298 40S ribosomal protein S11; K02949 small subunit ... 134 8e-32
mmu:27207 Rps11; ribosomal protein S11; K02949 small subunit r... 114 8e-26
hsa:6205 RPS11; ribosomal protein S11; K02949 small subunit ri... 114 8e-26
dre:406686 rps11, wu:fa91c09, wu:fb34b11, zgc:56293; ribosomal... 114 1e-25
xla:380368 rps11, MGC64491; ribosomal protein S11; K02949 smal... 112 3e-25
sce:YDR025W RPS11A; Rps11ap; K02949 small subunit ribosomal pr... 108 6e-24
sce:YBR048W RPS11B; Rps11bp; K02949 small subunit ribosomal pr... 108 6e-24
ath:AT4G30800 40S ribosomal protein S11 (RPS11B); K02949 small... 105 3e-23
cel:F40F11.1 rps-11; Ribosomal Protein, Small subunit family m... 105 5e-23
ath:AT5G23740 RPS11-BETA (RIBOSOMAL PROTEIN S11-BETA); structu... 104 7e-23
ath:AT3G48930 EMB1080 (embryo defective 1080); structural cons... 103 1e-22
mmu:100039525 Gm9808; ribosomal protein S11 pseudogene 100 1e-21
eco:b3311 rpsQ, ECK3298, JW3273, neaA; 30S ribosomal subunit p... 35.4 0.054
hsa:219285 SAMD9L, C7orf6, DRIF2, FLJ39885, KIAA2005, UEF1; st... 35.0 0.071
dre:405866 MGC85942; zgc:85942 33.1 0.22
cel:C01C7.1 ark-1; A Ras-regulating Kinase family member (ark-... 32.0 0.62
dre:170846 appb, wu:fc43a03, zgc:92771; amyloid beta (A4) prec... 31.6 0.82
ath:AT3G10270 ATP binding / DNA binding / DNA topoisomerase (A... 29.3 3.3
ath:AT1G79850 RPS17; RPS17 (RIBOSOMAL PROTEIN S17); structural... 28.5 5.8
ath:AT1G49400 emb1129 (embryo defective 1129); structural cons... 28.5 5.8
> tgo:TGME49_026970 40S ribosomal protein S11, putative ; K02949
small subunit ribosomal protein S11e
Length=161
Score = 176 bits (447), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 99/157 (63%), Positives = 105/157 (66%), Gaps = 35/157 (22%)
Query 14 DVQTEKAFQRQHGASQLA---LAQSSKKGGR--RDTG----------------------G 46
DVQTE+AFQ+Q G S L L K+ R +D G G
Sbjct 5 DVQTERAFQKQDGVSSLGRLHLTGQKKRTQRYWKDVGLGFQTPRLAKESKYVDKKCPFTG 64
Query 47 SM--------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 98
++ GMVIS KMKR VVIRR YLHFV KYSR EKRHKNVTCHLSPCFEQVKEGD
Sbjct 65 NVSIRGRVIKGMVISTKMKRAVVIRRNYLHFVPKYSRFEKRHKNVTCHLSPCFEQVKEGD 124
Query 99 IVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
IVTAGQCRPLSKTIRFNVLKVE NQVFGNVRKQFRLF
Sbjct 125 IVTAGQCRPLSKTIRFNVLKVEKNQVFGNVRKQFRLF 161
> pfa:PFC0775w 40S ribosomal protein S11, putative; K02949 small
subunit ribosomal protein S11e
Length=161
Score = 147 bits (370), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 82/161 (50%), Positives = 99/161 (61%), Gaps = 36/161 (22%)
Query 10 ATQQDVQTEKAFQRQHGAS-----------------------QLALAQSSKKGGRRD--- 43
AT DVQ E+A+Q+Q GAS A + +K+G D
Sbjct 2 ATTLDVQHERAYQKQEGASFFNSKKIKKGSKSYIRYWKKVGLGFATPKEAKEGVYVDKKC 61
Query 44 --TGGSM-------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQV 94
TG GMVIS KMKR ++IRR YLH+V+KY+R EKRHKN+ CH SPCF+ V
Sbjct 62 PFTGNVSIRGRILKGMVISNKMKRTIIIRRNYLHYVKKYNRFEKRHKNIPCHCSPCFD-V 120
Query 95 KEGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
KEGDIVT GQCRPLSKT+RFNVL VE +Q+FG+ RKQF LF
Sbjct 121 KEGDIVTVGQCRPLSKTVRFNVLHVEKHQIFGSARKQFVLF 161
> bbo:BBOV_II004570 18.m06381; 40S ribosomal protein S11; K02949
small subunit ribosomal protein S11e
Length=156
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 75/153 (49%), Positives = 92/153 (60%), Gaps = 32/153 (20%)
Query 14 DVQTEKAFQRQHGASQLALAQSSKKG-GRRDTGGSM------------------------ 48
DVQ KA+Q+Q S L + +K +D G
Sbjct 5 DVQVHKAYQKQPNVSVLKFQKLGRKSRYYKDVGMGFETPKEAKEGTYIDKKCPFTGNVSI 64
Query 49 ------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTA 102
GMV+SAKMKR +++R +YLH+V+KY+R EKRH+N+ CH SPCFE GDIVT
Sbjct 65 RGRIIKGMVLSAKMKRTIIMRVSYLHYVKKYNRFEKRHRNIPCHCSPCFEPT-AGDIVTV 123
Query 103 GQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
GQCRPLSKT+RFNVLKVE NQ+FGN RKQFRLF
Sbjct 124 GQCRPLSKTVRFNVLKVEKNQIFGNPRKQFRLF 156
> tpv:TP02_0298 40S ribosomal protein S11; K02949 small subunit
ribosomal protein S11e
Length=156
Score = 134 bits (337), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 78/153 (50%), Positives = 90/153 (58%), Gaps = 32/153 (20%)
Query 14 DVQTEKAFQRQHGASQLAL-------------------AQSSKKGGRRD-----TGGSM- 48
DVQ KAFQ+Q S L + +K+G D TG
Sbjct 5 DVQVHKAFQKQANVSIKKLQKLGNKARYWKDVGMGFVTPKEAKEGHYVDKKCPFTGNVSI 64
Query 49 ------GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTA 102
GMVIS KMK +V+R YL +V KY+R EKRHKNV CH+SPCF V GDIVT
Sbjct 65 RGRVLKGMVISHKMKNTLVMRVNYLQYVPKYNRFEKRHKNVPCHVSPCFT-VSAGDIVTV 123
Query 103 GQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
GQCRPLSKT+RFNVLKVE N++FGN RKQFRLF
Sbjct 124 GQCRPLSKTVRFNVLKVEKNEIFGNPRKQFRLF 156
> mmu:27207 Rps11; ribosomal protein S11; K02949 small subunit
ribosomal protein S11e
Length=158
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 66/87 (75%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V+ GDIVT G+CRPL
Sbjct 75 GVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVQIGDIVTVGECRPL 134
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 135 SKTVRFNVLKVTKA---AGTKKQFQKF 158
> hsa:6205 RPS11; ribosomal protein S11; K02949 small subunit
ribosomal protein S11e
Length=158
Score = 114 bits (285), Expect = 8e-26, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 66/87 (75%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V+ GDIVT G+CRPL
Sbjct 75 GVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVQIGDIVTVGECRPL 134
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 135 SKTVRFNVLKVTKA---AGTKKQFQKF 158
> dre:406686 rps11, wu:fa91c09, wu:fb34b11, zgc:56293; ribosomal
protein S11; K02949 small subunit ribosomal protein S11e
Length=159
Score = 114 bits (284), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 53/87 (60%), Positives = 65/87 (74%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V GDIVT G+CRPL
Sbjct 76 GVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVTVGDIVTVGECRPL 135
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 136 SKTVRFNVLKVTKA---AGAKKQFQKF 159
> xla:380368 rps11, MGC64491; ribosomal protein S11; K02949 small
subunit ribosomal protein S11e
Length=158
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 52/87 (59%), Positives = 65/87 (74%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V+ GD VT G+CRPL
Sbjct 75 GVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVQVGDTVTVGECRPL 134
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 135 SKTVRFNVLKVTKA---AGTKKQFQKF 158
> sce:YDR025W RPS11A; Rps11ap; K02949 small subunit ribosomal
protein S11e
Length=156
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 84/158 (53%), Gaps = 35/158 (22%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKGGRRDTGGSMGM------------------- 50
+T+ VQ+E+AFQ+Q ++SK+ R +G
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKRTKRWYKNAGLGFKTPKTAIEGSYIDKKCPFT 61
Query 51 -------------VISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEG 97
V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+ G
Sbjct 62 GLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQVG 120
Query 98 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
DIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 121 DIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> sce:YBR048W RPS11B; Rps11bp; K02949 small subunit ribosomal
protein S11e
Length=156
Score = 108 bits (269), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 63/158 (39%), Positives = 84/158 (53%), Gaps = 35/158 (22%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKGGRRDTGGSMGM------------------- 50
+T+ VQ+E+AFQ+Q ++SK+ R +G
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKRTKRWYKNAGLGFKTPKTAIEGSYIDKKCPFT 61
Query 51 -------------VISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEG 97
V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+ G
Sbjct 62 GLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQVG 120
Query 98 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 135
DIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 121 DIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> ath:AT4G30800 40S ribosomal protein S11 (RPS11B); K02949 small
subunit ribosomal protein S11e
Length=159
Score = 105 bits (263), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 49/71 (69%), Positives = 58/71 (81%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD VT GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRDYLHFVKKYRRYEKRHSNIPAHVSPCF-RVKEGDRVTIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> cel:F40F11.1 rps-11; Ribosomal Protein, Small subunit family
member (rps-11); K02949 small subunit ribosomal protein S11e
Length=155
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 48/87 (55%), Positives = 61/87 (70%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V+ KM R +V+RR YLH+++KY R EKRHKNV H SP F + GD+VT G+CRPL
Sbjct 72 GVVLKNKMTRTIVVRRDYLHYIKKYRRYEKRHKNVPAHCSPAFRDIHPGDLVTIGECRPL 131
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV + G +K F F
Sbjct 132 SKTVRFNVLKVNKS---GTSKKGFSKF 155
> ath:AT5G23740 RPS11-BETA (RIBOSOMAL PROTEIN S11-BETA); structural
constituent of ribosome; K02949 small subunit ribosomal
protein S11e
Length=159
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 48/71 (67%), Positives = 57/71 (80%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD V GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRNYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGDHVIIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> ath:AT3G48930 EMB1080 (embryo defective 1080); structural constituent
of ribosome; K02949 small subunit ribosomal protein
S11e
Length=160
Score = 103 bits (258), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 47/71 (66%), Positives = 57/71 (80%), Gaps = 1/71 (1%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD + GQCRPL
Sbjct 74 GTCHSAKMQRTIIVRRDYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGDHIIIGQCRPL 132
Query 109 SKTIRFNVLKV 119
SKT+RFNVLKV
Sbjct 133 SKTVRFNVLKV 143
> mmu:100039525 Gm9808; ribosomal protein S11 pseudogene
Length=158
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 49/87 (56%), Positives = 63/87 (72%), Gaps = 3/87 (3%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G+V KM+R +VI + YL ++RKY+R EK HKN++ HLSPCF V+ GDIVT G+CRPL
Sbjct 75 GVVTKMKMQRTIVICQDYLCYIRKYNRFEKCHKNMSVHLSPCFRDVQIGDIVTVGECRPL 134
Query 109 SKTIRFNVLKVESNQVFGNVRKQFRLF 135
SKT+RFNVLKV +KQF+ F
Sbjct 135 SKTVRFNVLKVTKA---AGTKKQFQKF 158
> eco:b3311 rpsQ, ECK3298, JW3273, neaA; 30S ribosomal subunit
protein S17; K02961 small subunit ribosomal protein S17
Length=84
Score = 35.4 bits (80), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 22/84 (26%), Positives = 41/84 (48%), Gaps = 16/84 (19%)
Query 49 GMVISAKMKRCVVI-------RRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVT 101
G V+S KM++ +V+ Y F+++ +++ +N C + GD+V
Sbjct 10 GRVVSDKMEKSIVVAIERFVKHPIYGKFIKRTTKLHVHDENNECGI---------GDVVE 60
Query 102 AGQCRPLSKTIRFNVLKVESNQVF 125
+CRPLSKT + +++V V
Sbjct 61 IRECRPLSKTKSWTLVRVVEKAVL 84
> hsa:219285 SAMD9L, C7orf6, DRIF2, FLJ39885, KIAA2005, UEF1;
sterile alpha motif domain containing 9-like
Length=1584
Score = 35.0 bits (79), Expect = 0.071, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 71 RKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPLSKTIR 113
+K SR +++ + CHL PC Q KE ++ CR + +R
Sbjct 1294 KKVSRCFRKYTELFCHLDPCLLQSKESQLLQEENCRKKLEALR 1336
> dre:405866 MGC85942; zgc:85942
Length=363
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 20/44 (45%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query 33 AQSSKKGGRRDTGGSMGMVIS---AKMKRCVVIRRTYLHFVRKY 73
+ SS GRRD GGS G V S A+ +RRTY+ V KY
Sbjct 15 SNSSAASGRRDPGGSSGRVTSDLRARPLPPACLRRTYMEGVFKY 58
> cel:C01C7.1 ark-1; A Ras-regulating Kinase family member (ark-1);
K08252 receptor protein-tyrosine kinase [EC:2.7.10.1]
Length=1043
Score = 32.0 bits (71), Expect = 0.62, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 91 FEQVKEGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFR 133
+ V+E D+++ G RP K +R K+ + +FG V+K F+
Sbjct 42 LQYVEEVDLLSVGMSRPEQKRLRKEYTKMFPSGIFGKVKKAFK 84
> dre:170846 appb, wu:fc43a03, zgc:92771; amyloid beta (A4) precursor
protein b; K04520 amyloid beta (A4) protein
Length=694
Score = 31.6 bits (70), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 32/67 (47%), Gaps = 4/67 (5%)
Query 64 RTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPLSKTIRFNVLKVESNQ 123
R L+ ++KY R E++ + H FE V+E D A Q RP T V++ NQ
Sbjct 428 RQVLNLLKKYIRAEQKDRQ---HTLKHFEHVREVDPKKASQIRPFVMT-HLRVIEERMNQ 483
Query 124 VFGNVRK 130
G + K
Sbjct 484 SLGYLYK 490
> ath:AT3G10270 ATP binding / DNA binding / DNA topoisomerase
(ATP-hydrolyzing); K02470 DNA gyrase subunit B [EC:5.99.1.3]
Length=693
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 36 SKKGGRRDTGGSMGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSP 89
S GG G S+ +S ++ V++RR + F +KYSR K +TCH+ P
Sbjct 123 SVSGGLHGVGLSVVNALSEALE--VIVRRDGMEFQQKYSR-GKPVTTLTCHVLP 173
> ath:AT1G79850 RPS17; RPS17 (RIBOSOMAL PROTEIN S17); structural
constituent of ribosome; K02961 small subunit ribosomal protein
S17
Length=149
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 31/71 (43%), Gaps = 2/71 (2%)
Query 49 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 108
G V+ A + V + L KY R + K H P Q K GD+V + RP+
Sbjct 55 GRVVCATSDKTVAVEVVRLAPHPKYKRRVRMKKKYQAH-DPD-NQFKVGDVVRLEKSRPI 112
Query 109 SKTIRFNVLKV 119
SKT F L V
Sbjct 113 SKTKSFVALPV 123
> ath:AT1G49400 emb1129 (embryo defective 1129); structural constituent
of ribosome; K02961 small subunit ribosomal protein
S17
Length=116
Score = 28.5 bits (62), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query 48 MGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRP 107
+G V+S KM++ VV+ L + Y+R KR H + GD V RP
Sbjct 5 IGTVVSNKMQKSVVVAVDRLFHNKIYNRYVKRTSKFMAHDD--KDACNIGDRVKLDPSRP 62
Query 108 LSKTIRFNVLKV 119
LSK + V ++
Sbjct 63 LSKNKHWIVAEI 74
Lambda K H
0.322 0.132 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2296762580
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40