bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2479_orf1
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
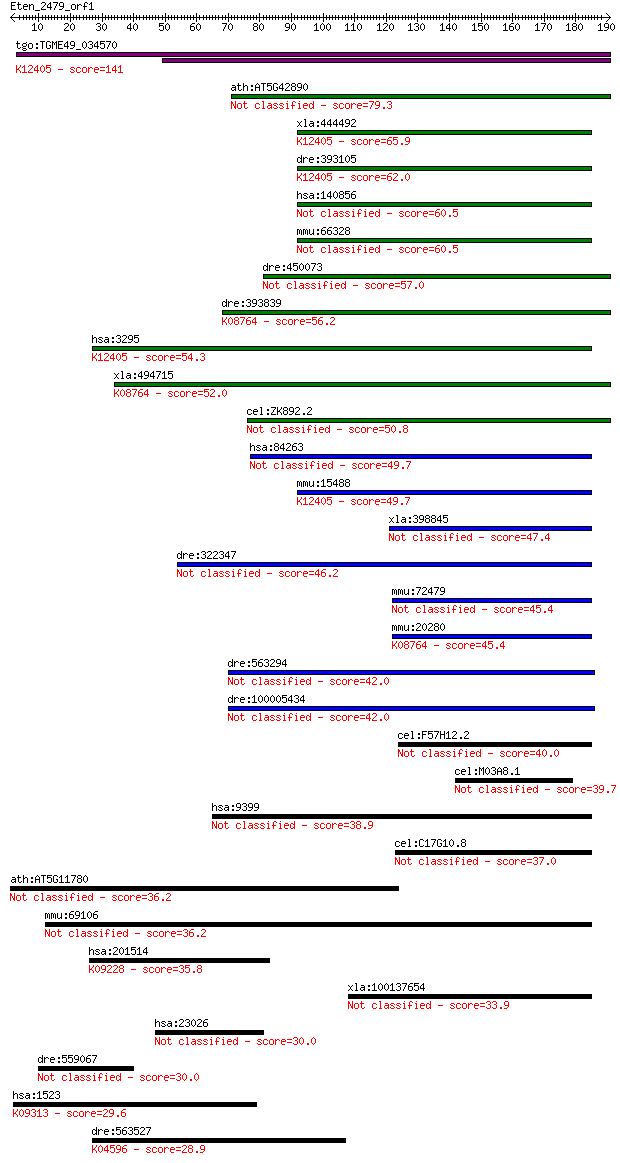
tgo:TGME49_034570 peroxisomal multifunctional enzyme type 2, p... 141 1e-33
ath:AT5G42890 SCP2; SCP2 (STEROL CARRIER PROTEIN 2); oxidoredu... 79.3 7e-15
xla:444492 hsd17b4, MGC81885; hydroxysteroid (17-beta) dehydro... 65.9 8e-11
dre:393105 hsd17b4, MGC55545, zgc:55545, zgc:77300; hydroxyste... 62.0 1e-09
hsa:140856 C20orf79, HSD22, MGC138229, dJ1068E13.2; chromosome... 60.5 4e-09
mmu:66328 1700010M22Rik; RIKEN cDNA 1700010M22 gene 60.5 4e-09
dre:450073 zgc:101621 57.0 4e-08
dre:393839 scp2, MGC77634, zgc:77634; sterol carrier protein 2... 56.2 6e-08
hsa:3295 HSD17B4, DBP, MFE-2, MPF-2, SDR8C1; hydroxysteroid (1... 54.3 3e-07
xla:494715 scp2; sterol carrier protein 2 (EC:2.3.1.176); K087... 52.0 1e-06
cel:ZK892.2 nlt-1; Non-specific Lipid Transfer protein family ... 50.8 3e-06
hsa:84263 HSDL2, C9orf99, FLJ25855, MGC10940, SDR13C1; hydroxy... 49.7 6e-06
mmu:15488 Hsd17b4, 17-beta-HSD, 17[b]-HSD, 4, DBP, MFE-2, MFP2... 49.7 7e-06
xla:398845 hsdl2, MGC68498; hydroxysteroid dehydrogenase like 2 47.4
dre:322347 hsdl2, wu:fb58f08, zgc:73140, zgc:77693; hydroxyste... 46.2 6e-05
mmu:72479 Hsdl2, 2610207I16Rik; hydroxysteroid dehydrogenase l... 45.4 1e-04
mmu:20280 Scp2, AA409774, AA409893, C76618, C79031, NSL-TP, SC... 45.4 1e-04
dre:563294 im:7145864; wu:fd21f07 42.0 0.001
dre:100005434 zgc:158861 42.0 0.001
cel:F57H12.2 unc-24; UNCoordinated family member (unc-24) 40.0 0.005
cel:M03A8.1 dhs-28; DeHydrogenases, Short chain family member ... 39.7 0.006
hsa:9399 STOML1, FLJ36370, SLP-1, STORP, hUNC-24; stomatin (EP... 38.9 0.012
cel:C17G10.8 dhs-6; DeHydrogenases, Short chain family member ... 37.0 0.036
ath:AT5G11780 hypothetical protein 36.2 0.062
mmu:69106 Stoml1, 1810015E19Rik, AI847431, MGC13700, SLP1, STO... 36.2 0.067
hsa:201514 ZNF584, FLJ39899; zinc finger protein 584; K09228 K... 35.8 0.088
xla:100137654 stoml1; stomatin (EPB72)-like 1 33.9
hsa:23026 MYO16, KIAA0865, MYR8, Myo16b; myosin XVI 30.0 4.5
dre:559067 si:dkeyp-77c8.1, si:dkeyp-77c8.2; si:dkeyp-77c8.3 30.0 4.7
hsa:1523 CUX1, CASP, CDP, CDP/Cut, CDP1, COY1, CUTL1, CUX, Clo... 29.6 5.9
dre:563527 brain-specific angiogenesis inhibitor 3-like; K0459... 28.9 9.7
> tgo:TGME49_034570 peroxisomal multifunctional enzyme type 2,
putative (EC:2.3.1.176 4.2.1.107); K12405 3-hydroxyacyl-CoA
dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35 4.2.1.107]
Length=625
Score = 141 bits (356), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 77/200 (38%), Positives = 118/200 (59%), Gaps = 18/200 (9%)
Query 3 GKTFPSSFTLEDLSKEWKTVLDFKDKVSYPSSLQDSLIMASQQI-----------VSEGK 51
G +F + + +D+++EW+ + DF +V+YP+SLQDS++M QQ+ G
Sbjct 252 GHSFATPLSPDDIAREWRHIRDFSGEVAYPASLQDSMLMVMQQLQQGTKPTDKRAEQRGA 311
Query 52 AAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIA 111
+ A E+ + A A +AA G KAE IFK++ AY+ + Q+L+ KV+S F F +
Sbjct 312 SDAKEEKKSGASAQEGSAAIG---KAEVIFKLMDAYIRNAPDARQKLQEKVDSVFGFNVT 368
Query 112 AKQQQQPVLQWTIDLRQ-DGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQG 170
+ + W+I+L++ + G + G+ + D F + E F +CLG LNPQ+AF+QG
Sbjct 369 DGKTTK---SWSINLKKGNAGGGVEDGLHSSPDVLFTMAAENFVSVCLGTLNPQMAFVQG 425
Query 171 KMKIKGSMAAAAKFTPQLFP 190
KMKIKGSM A KFTP LFP
Sbjct 426 KMKIKGSMQKATKFTPSLFP 445
Score = 70.9 bits (172), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 46/144 (31%), Positives = 76/144 (52%), Gaps = 10/144 (6%)
Query 49 EGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLF 108
E K A G A P+P A K+ +++++ +L + ++L K+ S +
Sbjct 485 EEKDAKHGSGKAQVPSPRAKQL-----KSMTLYEVMKRHLA--TPEGEKLVKKIKSVYRL 537
Query 109 EIAAKQQQQPVLQWTIDLRQDGEGCAKAGV--WGAADATFAVGEEAFCRICLGELNPQVA 166
I ++ PV + +DL+ + A D T + +E F ++ LG++NPQ+A
Sbjct 538 NILPRKGADPV-KVILDLKNMPPSIREEAEQEHAACDCTITLLDEDFVKLALGKMNPQLA 596
Query 167 FLQGKMKIKGSMAAAAKFTPQLFP 190
F+QGK+K+KGSM AA KFTP +FP
Sbjct 597 FVQGKIKLKGSMQAALKFTPDIFP 620
> ath:AT5G42890 SCP2; SCP2 (STEROL CARRIER PROTEIN 2); oxidoreductase/
sterol carrier
Length=123
Score = 79.3 bits (194), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 42/120 (35%), Positives = 70/120 (58%), Gaps = 3/120 (2%)
Query 71 AGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDG 130
A + K++ I M+ +L D K ++ K+ + IA K+ + + +DL++ G
Sbjct 2 ANTQLKSDAIMDMMKEHLSTDAGK--EVTEKIGLVYQINIAPKKLGFEEVTYIVDLKK-G 58
Query 131 EGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFTPQLFP 190
E G DATF+ ++ F ++ G++NPQ+AF++G MKIKGS++AA KFTP +FP
Sbjct 59 EVTKGKYEGGKVDATFSFKDDDFVKVATGKMNPQMAFIRGAMKIKGSLSAAQKFTPDIFP 118
> xla:444492 hsd17b4, MGC81885; hydroxysteroid (17-beta) dehydrogenase
4; K12405 3-hydroxyacyl-CoA dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35
4.2.1.107]
Length=741
Score = 65.9 bits (159), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 54/94 (57%), Gaps = 3/94 (3%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAG-VWGAADATFAVGE 150
K+ QL KVN+ F ++I + + QWTIDL+ G G G G AD +F + +
Sbjct 640 KDVGGQLVKKVNAVFQWDIT--KDGKTASQWTIDLKSGGSGEVYRGKARGRADTSFTLSD 697
Query 151 EAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
E F + LG++NPQ AF GK+K+KG++ + K
Sbjct 698 EDFMELVLGKVNPQKAFFAGKLKVKGNIMLSQKL 731
> dre:393105 hsd17b4, MGC55545, zgc:55545, zgc:77300; hydroxysteroid
(17-beta) dehydrogenase 4; K12405 3-hydroxyacyl-CoA dehydrogenase
/ 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35 4.2.1.107]
Length=725
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/93 (35%), Positives = 53/93 (56%), Gaps = 4/93 (4%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEE 151
K+ ++L KVN+ F +EI + + WT+DL+ G G + AD TF V ++
Sbjct 627 KDSGEELVKKVNAVFGWEITTDGETRR--HWTVDLKT-GRGSVQRAA-AKADVTFTVSDQ 682
Query 152 AFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
F + +G+LNPQ AF GK+K+KG++ + K
Sbjct 683 DFMEVVMGKLNPQKAFFAGKLKVKGNIMLSQKL 715
> hsa:140856 C20orf79, HSD22, MGC138229, dJ1068E13.2; chromosome
20 open reading frame 79
Length=156
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 35/94 (37%), Positives = 51/94 (54%), Gaps = 4/94 (4%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAG-VWGAADATFAVGE 150
+E QL KVN+ F +I + + +L+WTIDL+ +G G G AD F + E
Sbjct 56 REVGAQLVKKVNAVFQLDIT--KNGKTILRWTIDLK-NGSGDMYPGPARLPADTVFTIPE 112
Query 151 EAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
F + LG++NPQ AFL GK K+ G + + K
Sbjct 113 SVFMELVLGKMNPQKAFLAGKFKVSGKVLLSWKL 146
> mmu:66328 1700010M22Rik; RIKEN cDNA 1700010M22 gene
Length=156
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 48/93 (51%), Gaps = 2/93 (2%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEE 151
KE QL KVN+ F +I + + +LQWTIDL+ AD F + +
Sbjct 56 KEVGAQLVKKVNAIFQLDIT--KDGKTILQWTIDLKNGAGDMYLGSARLPADTVFIIPDS 113
Query 152 AFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
F + +G++NPQ AFL GK K++G + + K
Sbjct 114 VFTELVVGKINPQKAFLAGKFKVRGKVLLSQKL 146
> dre:450073 zgc:101621
Length=142
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 36/114 (31%), Positives = 58/114 (50%), Gaps = 6/114 (5%)
Query 81 FKMLAAYLELDK---EKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAG 137
FK A + E++K E +Q K+ F F++ + + + W +D++ +G+G
Sbjct 26 FKAHAVFQEINKKLQEDGEQFVKKIGGVFAFKVKDGPEGKEAV-WIVDVK-NGKGSVHND 83
Query 138 VWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF-TPQLFP 190
AD T A+ + + G++NPQ AF QGK+KI G+M A K QL P
Sbjct 84 SDKKADCTIAMADSDLLDLMTGKMNPQTAFFQGKLKITGNMGMAMKLQNLQLQP 137
> dre:393839 scp2, MGC77634, zgc:77634; sterol carrier protein
2 (EC:2.3.1.176); K08764 sterol carrier protein 2 [EC:2.3.1.176]
Length=538
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 61/127 (48%), Gaps = 6/127 (4%)
Query 68 AAAAGSCSKAEKIFKMLAAYLELDK---EKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTI 124
AA + S S FK + E++K E+ + K+ F F++ L W +
Sbjct 409 AAVSTSASSDRSGFKAHTVFKEIEKKLQEEGEAYVKKIGGVFAFKVKDGPGGSDAL-WVV 467
Query 125 DLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
D++ +G+G + AD T A+ + + G+LNPQ AF QGK+KI G+M A K
Sbjct 468 DVK-NGKGSVSSDAGKKADCTIAMADADLLDMMTGQLNPQTAFFQGKLKITGNMGMAMKL 526
Query 185 -TPQLFP 190
QL P
Sbjct 527 QNLQLTP 533
> hsa:3295 HSD17B4, DBP, MFE-2, MPF-2, SDR8C1; hydroxysteroid
(17-beta) dehydrogenase 4 (EC:4.2.1.107 4.2.1.119); K12405 3-hydroxyacyl-CoA
dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35 4.2.1.107]
Length=736
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 38/158 (24%), Positives = 72/158 (45%), Gaps = 13/158 (8%)
Query 27 DKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAA 86
+++ + + +Q++ ++S + G +A P+ S E+I + L
Sbjct 582 NRIHFQTKVQET----GDIVISNAYVDLAPTSGTSAKTPSEGGKLQSTFVFEEIGRRL-- 635
Query 87 YLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATF 146
K+ ++ KVN+ F + I +WTIDL+ + GAAD T
Sbjct 636 -----KDIGPEVVKKVNAVFEWHITKGGNIGA--KWTIDLKSGSGKVYQGPAKGAADTTI 688
Query 147 AVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
+ +E F + LG+L+PQ AF G++K +G++ + K
Sbjct 689 ILSDEDFMEVVLGKLDPQKAFFSGRLKARGNIMLSQKL 726
> xla:494715 scp2; sterol carrier protein 2 (EC:2.3.1.176); K08764
sterol carrier protein 2 [EC:2.3.1.176]
Length=536
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/158 (27%), Positives = 72/158 (45%), Gaps = 6/158 (3%)
Query 34 SLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAAYLELDKE 93
+LQ ++ + +VS K ++ A +A KA+ +FK + L E
Sbjct 379 ALQHNIGLGGAVVVSIYKMGFPDSANKSSGIRMVATSASDGFKAQFVFKEIEQKLA---E 435
Query 94 KTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEEAF 153
+QL K+ F F++ + W +D++ +G+G AD T + +
Sbjct 436 GGEQLVKKIGGIFAFKVKDGPGGKEA-TWVVDVK-NGKGSVDVNSDKKADCTITMADADL 493
Query 154 CRICLGELNPQVAFLQGKMKIKGSMAAAAKF-TPQLFP 190
+ G++NPQ AF QGK+KI G+M A K + QL P
Sbjct 494 LALMTGKMNPQTAFFQGKLKIAGNMGMAMKLQSLQLQP 531
> cel:ZK892.2 nlt-1; Non-specific Lipid Transfer protein family
member (nlt-1)
Length=118
Score = 50.8 bits (120), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/119 (30%), Positives = 57/119 (47%), Gaps = 15/119 (12%)
Query 76 KAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQD----GE 131
K++ IF+ + + DKE + KV + F IA + V WTID + D G+
Sbjct 4 KSDVIFEEIKERIATDKEMVK----KVGTSFRMTIAGADGKTKV--WTIDAKSDTPYVGD 57
Query 132 GCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFTPQLFP 190
++ + + + F I G++ P AF+QGKMK+KG++A A K L P
Sbjct 58 DSSRP-----VEIEINIKDSDFIAIAAGKMKPDQAFMQGKMKLKGNIAKAMKLRTILDP 111
> hsa:84263 HSDL2, C9orf99, FLJ25855, MGC10940, SDR13C1; hydroxysteroid
dehydrogenase like 2
Length=345
Score = 49.7 bits (117), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 52/108 (48%), Gaps = 10/108 (9%)
Query 77 AEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKA 136
E+ F+++ L D K Q + +LFE++ + W +DL+ G
Sbjct 237 VEETFRIVKDSLSDDVVKATQ------AIYLFELSGEDGGT----WFLDLKSKGGNVGYG 286
Query 137 GVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
AD ++ + F ++ G+L P +AF+ GK+KIKG+MA A K
Sbjct 287 EPSDQADVVMSMTTDDFVKMFSGKLKPTMAFMSGKLKIKGNMALAIKL 334
> mmu:15488 Hsd17b4, 17-beta-HSD, 17[b]-HSD, 4, DBP, MFE-2, MFP2,
MPF-2, Mfp-2, perMFE-2; hydroxysteroid (17-beta) dehydrogenase
4 (EC:4.2.1.107 4.2.1.119); K12405 3-hydroxyacyl-CoA
dehydrogenase / 3a,7a,12a-trihydroxy-5b-cholest-24-enoyl-CoA
hydratase [EC:1.1.1.35 4.2.1.107]
Length=735
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 47/93 (50%), Gaps = 2/93 (2%)
Query 92 KEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEE 151
K +++ K N+ F + I + +WTIDL+ + G+AD T + +E
Sbjct 635 KSVGREVVKKANAVFEWHIT--KGGTVAAKWTIDLKSGSGEVYQGPAKGSADVTIIISDE 692
Query 152 AFCRICLGELNPQVAFLQGKMKIKGSMAAAAKF 184
F + G+L+PQ AF G++K +G++ + K
Sbjct 693 DFMEVVFGKLDPQKAFFSGRLKARGNIMLSQKL 725
> xla:398845 hsdl2, MGC68498; hydroxysteroid dehydrogenase like
2
Length=417
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 121 QWTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAA 180
W +DL+ D G K AD ++ F ++ G++ P +AF+ GK+KIKG M
Sbjct 343 NWFLDLKNDKGGVGKGEPSTKADVVMSMDSGDFIKMFAGKMKPTMAFMSGKLKIKGDMGL 402
Query 181 AAKF 184
A K
Sbjct 403 ALKL 406
> dre:322347 hsdl2, wu:fb58f08, zgc:73140, zgc:77693; hydroxysteroid
dehydrogenase like 2
Length=415
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 36/131 (27%), Positives = 54/131 (41%), Gaps = 13/131 (9%)
Query 54 ASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAK 113
A G A A A AAG S+ +F + + + KT Q + F +A +
Sbjct 287 AHGATPAFTTAKADPVAAGPVSE---MFNTIRGIISPEMVKTTQ------GVYKFNLAGE 337
Query 114 QQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMK 173
W +DL+ D AD ++ E F ++ G+L P +AF+ GK+
Sbjct 338 HAGV----WYLDLKNDAGSAGNGEPPVKADVVMSMDSEDFVKMFGGKLKPTMAFMSGKLT 393
Query 174 IKGSMAAAAKF 184
IKG M A K
Sbjct 394 IKGDMGLAIKL 404
> mmu:72479 Hsdl2, 2610207I16Rik; hydroxysteroid dehydrogenase
like 2
Length=490
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 122 WTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAA 181
W +DL+ G AD ++ + F ++ G+L P +AF+ GK+KIKG++A A
Sbjct 417 WFLDLKSKGGKVGHGEPSDRADVVMSMATDDFVKMFSGKLKPTMAFMSGKLKIKGNIALA 476
Query 182 AKF 184
K
Sbjct 477 IKL 479
> mmu:20280 Scp2, AA409774, AA409893, C76618, C79031, NSL-TP,
SCP-2, SCPx, ns-LTP; sterol carrier protein 2, liver (EC:2.3.1.176);
K08764 sterol carrier protein 2 [EC:2.3.1.176]
Length=547
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 34/63 (53%), Gaps = 1/63 (1%)
Query 122 WTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAA 181
W +D++ +G+G AD T + + + G++NPQ AF QGK+KI G+M A
Sbjct 474 WVVDVK-NGKGSVLPNSDKKADCTITMADSDLLALMTGKMNPQSAFFQGKLKIAGNMGLA 532
Query 182 AKF 184
K
Sbjct 533 MKL 535
> dre:563294 im:7145864; wu:fd21f07
Length=410
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 52/117 (44%), Gaps = 10/117 (8%)
Query 70 AAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQD 129
A S S ++ + + + L +++L +V +CF F I Q + +DL Q
Sbjct 294 AVSSVSSVNELIETVRSVL------SEELVHQVGACFHFHITTNSGQTS--SYYVDLTQ- 344
Query 130 GEGCAKAGVW-GAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFT 185
G G AGV D + + E+ + G L P A+ G+++++G + A K
Sbjct 345 GRGACGAGVLQREPDVSLCMSEQDLLAMFQGSLQPFAAYSSGRLRVQGDLNTAMKLN 401
> dre:100005434 zgc:158861
Length=410
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 29/117 (24%), Positives = 52/117 (44%), Gaps = 10/117 (8%)
Query 70 AAGSCSKAEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQWTIDLRQD 129
A S S ++ + + + L +++L +V +CF F I Q + +DL Q
Sbjct 294 AVSSVSSVNELIETVRSVL------SEELVHQVGACFHFHITTNSGQTS--SYYVDLTQ- 344
Query 130 GEGCAKAGVW-GAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAKFT 185
G G AGV D + + E+ + G L P A+ G+++++G + A K
Sbjct 345 GRGACGAGVLQREPDVSLCMSEQDLLAMFQGSLQPFAAYSSGRLRVQGDLNTAMKLN 401
> cel:F57H12.2 unc-24; UNCoordinated family member (unc-24)
Length=415
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 124 IDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAAAK 183
IDL+ G G A G D F E F +I E++P ++ G +K+KGS+ A +
Sbjct 345 IDLKH-GSGSAYKGTSLNPDVVFETSLEVFGKILTKEVSPVTVYMNGNLKVKGSIQDAMQ 403
Query 184 F 184
Sbjct 404 L 404
> cel:M03A8.1 dhs-28; DeHydrogenases, Short chain family member
(dhs-28)
Length=436
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 25/37 (67%), Gaps = 0/37 (0%)
Query 142 ADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSM 178
A+AT V + F I G+LN Q AF+ GK+K+KG++
Sbjct 382 ANATVTVADSDFVDIAAGKLNAQKAFMSGKLKVKGNV 418
> hsa:9399 STOML1, FLJ36370, SLP-1, STORP, hUNC-24; stomatin (EPB72)-like
1
Length=398
Score = 38.9 bits (89), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 53/124 (42%), Gaps = 13/124 (10%)
Query 65 PAAAAAAGSCSK---AEKIFKMLAAYLELDKEKTQQLKTKVNSCFLFEIAAKQQQQPVLQ 121
PA A S K AE + L +L ++ L ++V +C+ F + Q
Sbjct 276 PAPQVGARSSPKQPLAEGLLTALQPFL------SEALVSQVGACYQFNVVLPSGTQSA-- 327
Query 122 WTIDLRQDGEGCAKAGVW-GAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAA 180
+ +DL G G GV G D + E + EL P A++ G++K+KG +A
Sbjct 328 YFLDL-TTGRGRVGHGVPDGIPDVVVEMAEADLRALLCRELRPLGAYMSGRLKVKGDLAM 386
Query 181 AAKF 184
A K
Sbjct 387 AMKL 390
> cel:C17G10.8 dhs-6; DeHydrogenases, Short chain family member
(dhs-6)
Length=418
Score = 37.0 bits (84), Expect = 0.036, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query 123 TIDLRQDGEG-CAKAGVWGAADATFAVGEEAFCRICLGELNPQVAFLQGKMKIKGSMAAA 181
T+DL+ +GEG G AD F + E F + G+L P A + K++I G M A
Sbjct 345 TLDLK-NGEGALTDKKASGKADVKFTLAPEHFAPLFTGKLRPTTALMTKKLQISGDMPGA 403
Query 182 AKF 184
K
Sbjct 404 MKL 406
> ath:AT5G11780 hypothetical protein
Length=504
Score = 36.2 bits (82), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 32/126 (25%), Positives = 51/126 (40%), Gaps = 8/126 (6%)
Query 1 DQGKTFPSSFTLEDLSKEWKTVLDFKDKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGA 60
D S F+ DLS+ W V + +D L+ + AS+ + E + + G
Sbjct 149 DDDVLMVSGFSFADLSRVW-VVDEVEDNCRVEDCLEVFMPFASEILRKEIDSESCGVGYL 207
Query 61 AAPAPAAAAAAGSCSKAEKIFKMLAAYLELDK---EKTQQLKTKVNSCFLFEIAAKQQQQ 117
A + CS+ F + ELDK E Q+ + +SCF F++ K +
Sbjct 208 AGIVASQVFLLSLCSR----FDLDLGRSELDKDLQESVLQMISGFHSCFFFDVILKMLLE 263
Query 118 PVLQWT 123
P L T
Sbjct 264 PYLHLT 269
> mmu:69106 Stoml1, 1810015E19Rik, AI847431, MGC13700, SLP1, STORP,
UNC24, WPB72; stomatin-like 1
Length=399
Score = 36.2 bits (82), Expect = 0.067, Method: Compositional matrix adjust.
Identities = 43/201 (21%), Positives = 79/201 (39%), Gaps = 38/201 (18%)
Query 12 LEDLSKEWKTVLDF------------KDKVSYPSSLQDSLIMASQQIVSEGKAAASGEEG 59
+ D+++ W +D +D ++ PS L +L + ++ +A G
Sbjct 201 INDVTRAWGLEVDRVELAVEAVLQPPQDSLTVPS-LDSTLQQLALHLLGGSMNSAVGHVP 259
Query 60 AAAP----------APAAAAAAGSCSK-----AEKIFKMLAAYLELDKEKTQQLKTKVNS 104
+ P PA+ A AG AE + L +L ++ L ++V +
Sbjct 260 SPGPDTLEMINEVEPPASLAGAGPEPSPKQPVAEGLLTALQPFL------SEALVSQVGA 313
Query 105 CFLFEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVW-GAADATFAVGEEAFCRICLGELNP 163
C+ F + Q + + +DL G+G G G D + E + EL P
Sbjct 314 CYQFNVILPSGTQSI--YFLDL-TTGQGRVGHGEPDGIPDVVVEMAEADLQALLSKELRP 370
Query 164 QVAFLQGKMKIKGSMAAAAKF 184
A++ G++K+KG +A K
Sbjct 371 LGAYMSGRLKVKGDLAVVMKL 391
> hsa:201514 ZNF584, FLJ39899; zinc finger protein 584; K09228
KRAB domain-containing zinc finger protein
Length=421
Score = 35.8 bits (81), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 26 KDKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFK 82
+D+ ++P L+ ++ Q SEGK E GAA P ++ AEK+FK
Sbjct 104 EDERAHPEHLKSYRVIQHQDTHSEGKPRRHTEHGAAFPPGSSCGQQQEVHVAEKLFK 160
> xla:100137654 stoml1; stomatin (EPB72)-like 1
Length=363
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 16/77 (20%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query 108 FEIAAKQQQQPVLQWTIDLRQDGEGCAKAGVWGAADATFAVGEEAFCRICLGELNPQVAF 167
+++ + ++ +DL+ G G GV D T + E + G+L+P A+
Sbjct 279 YQLYVTMHGGEISEYFLDLK-SGSGNCGWGVHPCPDVTLEMTEADLMSLICGDLHPLTAY 337
Query 168 LQGKMKIKGSMAAAAKF 184
G++++ G++ A +
Sbjct 338 TGGRLRVSGNIQTALQL 354
> hsa:23026 MYO16, KIAA0865, MYR8, Myo16b; myosin XVI
Length=1880
Score = 30.0 bits (66), Expect = 4.5, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 17/34 (50%), Gaps = 0/34 (0%)
Query 47 VSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKI 80
V+ GKA S E P P +A AG CS KI
Sbjct 1624 VNAGKAGPSAEAPKVHPKPNSAPVAGPCSSFPKI 1657
> dre:559067 si:dkeyp-77c8.1, si:dkeyp-77c8.2; si:dkeyp-77c8.3
Length=4421
Score = 30.0 bits (66), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 10 FTLEDLSKEWKTVLDFKDKVSYPSSLQDSL 39
TL+++ EWK VLD + + + LQD+L
Sbjct 541 HTLKNIEDEWKRVLDLAQLLKHQAELQDTL 570
> hsa:1523 CUX1, CASP, CDP, CDP/Cut, CDP1, COY1, CUTL1, CUX, Clox,
Cux/CDP, FLJ31745, GOLIM6, Nbla10317, p100, p110, p200,
p75; cut-like homeobox 1; K09313 homeobox protein cut-like
Length=1516
Score = 29.6 bits (65), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 10/83 (12%)
Query 2 QGKTFPSSFTLEDLSKEWKTVLDFKDKV------SYPSSLQDSLIMASQQIVSEGKAAAS 55
Q K +PS T+EDL+ T L+ K +Y S ++ L + Q S+G+A AS
Sbjct 1275 QQKPYPSPKTIEDLA----TQLNLKTSTVINWFHNYRSRIRRELFIEEIQAGSQGQAGAS 1330
Query 56 GEEGAAAPAPAAAAAAGSCSKAE 78
A + A ++ SC E
Sbjct 1331 DSPSARSGRAAPSSEGDSCDGVE 1353
> dre:563527 brain-specific angiogenesis inhibitor 3-like; K04596
brain-specific angiogenesis inhibitor 1
Length=716
Score = 28.9 bits (63), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 19/80 (23%), Positives = 35/80 (43%), Gaps = 0/80 (0%)
Query 27 DKVSYPSSLQDSLIMASQQIVSEGKAAASGEEGAAAPAPAAAAAAGSCSKAEKIFKMLAA 86
D+VS S ++S + Q I SE + + E+G + + + + S + + +
Sbjct 533 DRVSVSCSERESPVQTVQNISSESQMTNTCEQGDSGNSGMSKSETVSTLSMSSLERRKSR 592
Query 87 YLELDKEKTQQLKTKVNSCF 106
Y ELD EK + + F
Sbjct 593 YAELDFEKIMHTRKRHQDMF 612
Lambda K H
0.315 0.128 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40