bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2476_orf1
Length=122
Score E
Sequences producing significant alignments: (Bits) Value
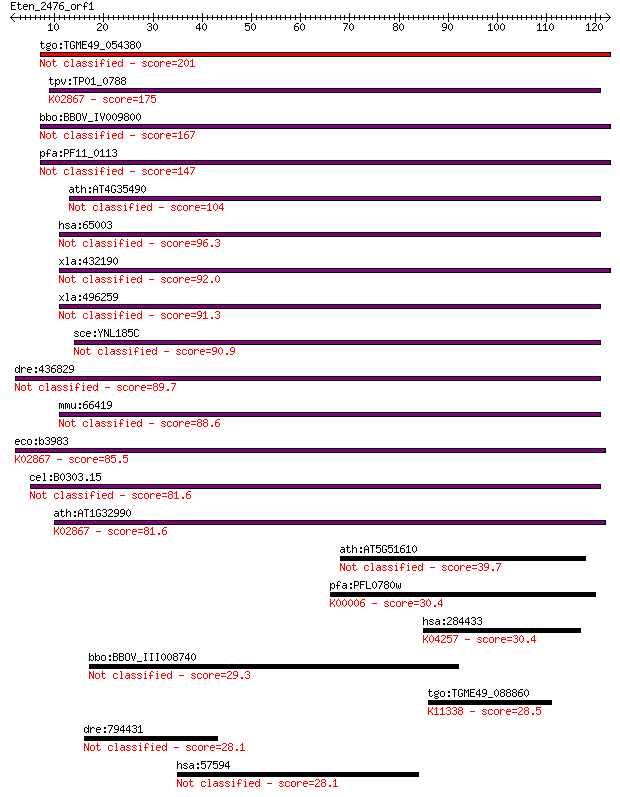
tgo:TGME49_054380 ribosomal protein L11, putative 201 3e-52
tpv:TP01_0788 60S ribosomal protein L11; K02867 large subunit ... 175 4e-44
bbo:BBOV_IV009800 23.m05843; ribosomal protein L11 167 7e-42
pfa:PF11_0113 mitochondrial ribosomal protein L11 precursor, p... 147 1e-35
ath:AT4G35490 MRPL11; MRPL11 (MITOCHONDRIAL RIBOSOMAL PROTEIN ... 104 7e-23
hsa:65003 MRPL11, L11MT, MGC111024, MRP-L11; mitochondrial rib... 96.3 2e-20
xla:432190 hypothetical protein MGC82344 92.0 4e-19
xla:496259 mrpl11; mitochondrial ribosomal protein L11 91.3 7e-19
sce:YNL185C MRPL19; Mrpl19p 90.9 9e-19
dre:436829 mrpl11, zgc:92770; mitochondrial ribosomal protein L11 89.7 2e-18
mmu:66419 Mrpl11, 2410001P07Rik, L11mt; mitochondrial ribosoma... 88.6 5e-18
eco:b3983 rplK, ECK3974, JW3946, relC; 50S ribosomal subunit p... 85.5 4e-17
cel:B0303.15 hypothetical protein 81.6 5e-16
ath:AT1G32990 PRPL11; PRPL11 (PLASTID RIBOSOMAL PROTEIN L11); ... 81.6 5e-16
ath:AT5G51610 ribosomal protein L11 family protein 39.7 0.003
pfa:PFL0780w glycerol-3-phosphate dehydrogenase, putative (EC:... 30.4 1.4
hsa:284433 OR10H5, OR19-25, OR19-26; olfactory receptor, famil... 30.4 1.5
bbo:BBOV_III008740 17.m10605; hypothetical protein 29.3 3.1
tgo:TGME49_088860 ruvB-like 2 protein, putative ; K11338 RuvB-... 28.5 5.2
dre:794431 v2rx1, si:dkey-183j10.1; vomeronasal 2 receptor, x1 28.1 7.4
hsa:57594 HOMEZ, KIAA1443; homeobox and leucine zipper encoding 28.1 7.4
> tgo:TGME49_054380 ribosomal protein L11, putative
Length=180
Score = 201 bits (512), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 90/116 (77%), Positives = 103/116 (88%), Gaps = 0/116 (0%)
Query 7 MTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPN 66
M+ +GRFRLIVPAA AKPSP+IGQTLGPLGINMM FCKEFNARTA +RP+VP+QVTIVP
Sbjct 1 MSVVGRFRLIVPAATAKPSPAIGQTLGPLGINMMQFCKEFNARTANVRPEVPLQVTIVPL 60
Query 67 ADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDWP 122
DR++KFS+R PS+ WFLLR ARCPMGSE G + +GNVTLKEIYHIA+CKSMDWP
Sbjct 61 TDRSYKFSIRAPSNIWFLLRTARCPMGSESPGHQSVGNVTLKEIYHIARCKSMDWP 116
> tpv:TP01_0788 60S ribosomal protein L11; K02867 large subunit
ribosomal protein L11
Length=177
Score = 175 bits (443), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 81/112 (72%), Positives = 91/112 (81%), Gaps = 0/112 (0%)
Query 9 NLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNAD 68
+GRFRLIVPAA AKPSPSIGQTLGPLGINM FCK+FN RT +IRP+VPVQV I + D
Sbjct 2 QIGRFRLIVPAAVAKPSPSIGQTLGPLGINMAEFCKKFNERTKEIRPNVPVQVLITTHHD 61
Query 69 RTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
++KFSLRTPSS WFL R AR PMGS E++GNVTLKE+YHIAKCK MD
Sbjct 62 NSYKFSLRTPSSQWFLRRIARVPMGSSNPKHEIVGNVTLKEVYHIAKCKCMD 113
> bbo:BBOV_IV009800 23.m05843; ribosomal protein L11
Length=178
Score = 167 bits (423), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 79/116 (68%), Positives = 91/116 (78%), Gaps = 0/116 (0%)
Query 7 MTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPN 66
M LGR+RLIVPAA AKPSPSIGQ LGPLGINM FCK+FN RT IRP+VP+QV I
Sbjct 1 MKFLGRYRLIVPAAVAKPSPSIGQALGPLGINMTDFCKKFNERTQLIRPNVPIQVRIYTL 60
Query 67 ADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDWP 122
++ ++KFSLRTP S WFL R AR PMGS + E++GNVTLKE+YHIAKCK MD P
Sbjct 61 SNASYKFSLRTPGSQWFLRRIARVPMGSSKPKHEIVGNVTLKEVYHIAKCKCMDPP 116
> pfa:PF11_0113 mitochondrial ribosomal protein L11 precursor,
putative
Length=178
Score = 147 bits (370), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 69/116 (59%), Positives = 84/116 (72%), Gaps = 0/116 (0%)
Query 7 MTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPN 66
M+ +GRF LIV + AKPS SIGQTLGPLGINMM F KEFN RT I +VP+QVT+ P
Sbjct 1 MSRIGRFNLIVLSGTAKPSASIGQTLGPLGINMMTFFKEFNDRTKCIAKNVPIQVTLEPL 60
Query 67 ADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDWP 122
DRT++F LRTP+ WF+ R AR PM S A +G++TL E++HIAKCK MD P
Sbjct 61 NDRTYRFYLRTPTVVWFIRRCARVPMFSSMAKHNTVGSITLAEVFHIAKCKRMDPP 116
> ath:AT4G35490 MRPL11; MRPL11 (MITOCHONDRIAL RIBOSOMAL PROTEIN
L11); structural constituent of ribosome
Length=155
Score = 104 bits (260), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 45/108 (41%), Positives = 71/108 (65%), Gaps = 0/108 (0%)
Query 13 FRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFK 72
RL VPA A+P+P +G LG +N+MAFCK+FNART K +PD P+ VTI D +F+
Sbjct 17 IRLTVPAGGARPAPPVGPALGQYRLNLMAFCKDFNARTQKYKPDTPMAVTITAFKDNSFE 76
Query 73 FSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
F++++P+ +W++ +AA GS + G + ++++ +Y IAK K D
Sbjct 77 FTVKSPTVSWYIKKAAGVDKGSTRPGHLSVTTLSVRHVYEIAKVKQTD 124
> hsa:65003 MRPL11, L11MT, MGC111024, MRP-L11; mitochondrial ribosomal
protein L11
Length=192
Score = 96.3 bits (238), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 65/110 (59%), Gaps = 0/110 (0%)
Query 11 GRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRT 70
G R IV A A P P +G LG G+++ FCKEFN RT I+ +P+ I+ DRT
Sbjct 18 GVIRAIVRAGLAMPGPPLGPVLGQRGVSINQFCKEFNERTKDIKEGIPLPTKILVKPDRT 77
Query 71 FKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
F+ + P+ ++FL AA G+ Q GKEV G VTLK +Y IA+ K+ D
Sbjct 78 FEIKIGQPTVSYFLKAAAGIEKGARQTGKEVAGLVTLKHVYEIARIKAQD 127
> xla:432190 hypothetical protein MGC82344
Length=192
Score = 92.0 bits (227), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 46/112 (41%), Positives = 63/112 (56%), Gaps = 0/112 (0%)
Query 11 GRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRT 70
G R +V A A P P +G LG GI + FCKEFN +T +++ +P+ V I DRT
Sbjct 18 GVIRALVRAGQAAPGPPLGPILGQRGIPIGQFCKEFNDKTKELKEGIPLPVKISVKPDRT 77
Query 71 FKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDWP 122
+ + P ++FL AA G+ Q G E+ G VTLK++Y IA KS D P
Sbjct 78 YDLKIGQPPVSYFLKAAAGIEKGASQTGHEIAGMVTLKQLYEIALVKSQDEP 129
> xla:496259 mrpl11; mitochondrial ribosomal protein L11
Length=192
Score = 91.3 bits (225), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 46/110 (41%), Positives = 61/110 (55%), Gaps = 0/110 (0%)
Query 11 GRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRT 70
G R +V A A P P +G LG GI + FCKEFN +T I+ +P+ V I DRT
Sbjct 18 GEIRALVRAGQAAPGPPLGPILGQRGIPIGQFCKEFNDKTKDIKEGIPLPVKIFVKPDRT 77
Query 71 FKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
+ + P ++FL AA G+ Q G E+ G VTLK++Y IA KS D
Sbjct 78 YDLKIGQPPVSYFLKAAAGIEKGASQTGHEIAGIVTLKQLYEIALVKSQD 127
> sce:YNL185C MRPL19; Mrpl19p
Length=158
Score = 90.9 bits (224), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 64/120 (53%), Gaps = 13/120 (10%)
Query 14 RLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKF 73
+LIV A A PSP +G LG GI + FCKEFNAR+A +P VPV V I DRTF F
Sbjct 11 KLIVGAGQAAPSPPVGPALGSKGIKAIDFCKEFNARSANYQPGVPVPVLITIKPDRTFTF 70
Query 74 SLRTPSSTWFLLRAARCPMGSEQAG-------------KEVIGNVTLKEIYHIAKCKSMD 120
+++P + + LL+A + G Q +G ++LK +Y IAK K D
Sbjct 71 EMKSPPTGYLLLKALKMDKGHGQPNVGTMLGSAPAKGPTRALGELSLKHVYEIAKIKKSD 130
> dre:436829 mrpl11, zgc:92770; mitochondrial ribosomal protein
L11
Length=196
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 45/119 (37%), Positives = 67/119 (56%), Gaps = 0/119 (0%)
Query 2 QAYMKMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQV 61
+A K G R IV + A P P +G LG GI + AFCK+FN RT ++ +P+ V
Sbjct 9 KAVKKSDTGGLIRAIVRSGQAAPGPPLGPILGQKGIPIGAFCKDFNERTKDLKEGIPLPV 68
Query 62 TIVPNADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
I DRT+ + P+ ++FL +AA G+ + G E+ G VT++ +Y IA+ KS D
Sbjct 69 KINVKPDRTYDLKIGQPTVSYFLKQAAGIEKGAGKTGHEIAGMVTVRAVYEIARVKSQD 127
> mmu:66419 Mrpl11, 2410001P07Rik, L11mt; mitochondrial ribosomal
protein L11
Length=192
Score = 88.6 bits (218), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 45/110 (40%), Positives = 62/110 (56%), Gaps = 0/110 (0%)
Query 11 GRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRT 70
G R IV A A P P +G LG G+++ FCKEFN +T I+ +P+ I DRT
Sbjct 18 GVIRSIVRAGQAIPGPPLGPILGQRGVSINQFCKEFNEKTKDIKEGIPLPTKIFIKPDRT 77
Query 71 FKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMD 120
F+ + P+ ++FL AA G+ GKEV G V+LK +Y IA K+ D
Sbjct 78 FELKIGQPTVSYFLKAAAGIEKGARHTGKEVAGLVSLKHVYEIACVKAKD 127
> eco:b3983 rplK, ECK3974, JW3946, relC; 50S ribosomal subunit
protein L11; K02867 large subunit ribosomal protein L11
Length=142
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 46/120 (38%), Positives = 66/120 (55%), Gaps = 8/120 (6%)
Query 2 QAYMKMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQV 61
QAY+K L V A A PSP +G LG G+N+M FCK FNA+T I +P+ V
Sbjct 6 QAYVK--------LQVAAGMANPSPPVGPALGQQGVNIMEFCKAFNAKTDSIEKGLPIPV 57
Query 62 TIVPNADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDW 121
I ADR+F F +TP + L +AA GS + K+ +G ++ ++ IA+ K+ D
Sbjct 58 VITVYADRSFTFVTKTPPAAVLLKKAAGIKSGSGKPNKDKVGKISRAQLQEIAQTKAADM 117
> cel:B0303.15 hypothetical protein
Length=195
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 44/117 (37%), Positives = 66/117 (56%), Gaps = 3/117 (2%)
Query 5 MKMTNLGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIV 64
+K+ + R + A A +P +G LG G+N+ FCKEFN T + VP+ I
Sbjct 15 VKVVHGALLRTNIKAQMASAAPPLGPQLGQRGLNVANFCKEFNKETGHFKQGVPLPTRIT 74
Query 65 PNADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGK-EVIGNVTLKEIYHIAKCKSMD 120
DRT+ + TP++TW L +AA +G +A K E++G +T+K +Y IAK KS D
Sbjct 75 VKPDRTYDLEICTPTTTWLLKQAA--GIGRGKASKDEIVGKLTVKHLYEIAKIKSRD 129
> ath:AT1G32990 PRPL11; PRPL11 (PLASTID RIBOSOMAL PROTEIN L11);
structural constituent of ribosome; K02867 large subunit ribosomal
protein L11
Length=222
Score = 81.6 bits (200), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 43/113 (38%), Positives = 70/113 (61%), Gaps = 3/113 (2%)
Query 10 LGRFRLIVPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTA-KIRPDVPVQVTIVPNAD 68
+G +L + A A P+P +G LG G+N+MAFCK++NARTA K +PV++T+ D
Sbjct 75 VGVIKLALEAGKATPAPPVGPALGSKGVNIMAFCKDYNARTADKAGYIIPVEITVF--DD 132
Query 69 RTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSMDW 121
++F F L+TP ++ LL+AA GS+ ++ +G +T+ ++ IA K D
Sbjct 133 KSFTFILKTPPASVLLLKAAGVEKGSKDPQQDKVGVITIDQLRTIAAEKLPDL 185
> ath:AT5G51610 ribosomal protein L11 family protein
Length=182
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 32/50 (64%), Gaps = 0/50 (0%)
Query 68 DRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCK 117
D++F F L+TP +++ LL+AA GS+ + +G +T+ ++ IA+ K
Sbjct 92 DQSFTFILKTPPTSFLLLKAAGVKKGSKDPKQIKVGMITIDQVRKIAEVK 141
> pfa:PFL0780w glycerol-3-phosphate dehydrogenase, putative (EC:1.1.1.8);
K00006 glycerol-3-phosphate dehydrogenase (NAD+)
[EC:1.1.1.8]
Length=367
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 27/54 (50%), Gaps = 6/54 (11%)
Query 66 NADRTFKFSLRTPSSTWFLLRAARCPMGSEQAGKEVIGNVTLKEIYHIAKCKSM 119
NA + +F TP TW L G+++ G VTLK +YH+ K K+M
Sbjct 283 NAKCSAEFIKSTPKKTWEELE------NEILKGQKLQGTVTLKYVYHMIKEKNM 330
> hsa:284433 OR10H5, OR19-25, OR19-26; olfactory receptor, family
10, subfamily H, member 5; K04257 olfactory receptor
Length=315
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 85 LRAARCPMGSEQAGKEVIGNVTLKEIYHIAKC 116
LR C +G AG V+G V I+H+A C
Sbjct 138 LRGCTCRVGCSWAGGLVMGMVVTSAIFHLAFC 169
> bbo:BBOV_III008740 17.m10605; hypothetical protein
Length=703
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 34/75 (45%), Gaps = 3/75 (4%)
Query 17 VPAANAKPSPSIGQTLGPLGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKFSLR 76
+P+ + +P+ S + LG G +A+ VP+ I+PN D+ +SL
Sbjct 254 IPSDDNEPTHSTHEGLGVKGSPSALIESHLHAQRMS---GVPLSPKIIPNEDKPMPYSLP 310
Query 77 TPSSTWFLLRAARCP 91
+ T F+L + P
Sbjct 311 SVQDTSFILPTEKIP 325
> tgo:TGME49_088860 ruvB-like 2 protein, putative ; K11338 RuvB-like
protein 2 [EC:3.6.4.12]
Length=508
Score = 28.5 bits (62), Expect = 5.2, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 86 RAARCPMGSEQAGKEVIGNVTLKEI 110
R +CP G Q KEV+ +VTL EI
Sbjct 231 RFVQCPEGELQKRKEVVHSVTLHEI 255
> dre:794431 v2rx1, si:dkey-183j10.1; vomeronasal 2 receptor,
x1
Length=868
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 9/27 (33%), Positives = 20/27 (74%), Gaps = 0/27 (0%)
Query 16 IVPAANAKPSPSIGQTLGPLGINMMAF 42
++ A++ P+ ++ QTLGP GI ++++
Sbjct 155 VIGLASSSPTRAVAQTLGPFGIPLISY 181
> hsa:57594 HOMEZ, KIAA1443; homeobox and leucine zipper encoding
Length=550
Score = 28.1 bits (61), Expect = 7.4, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 23/49 (46%), Gaps = 0/49 (0%)
Query 35 LGINMMAFCKEFNARTAKIRPDVPVQVTIVPNADRTFKFSLRTPSSTWF 83
+GI K + K+ P+ P Q+ +P + + K SL TP S F
Sbjct 165 IGIGPPTLSKPTQTKGLKVEPEEPSQMPPLPQSHQKLKESLMTPGSGAF 213
Lambda K H
0.322 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2008132680
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40