bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
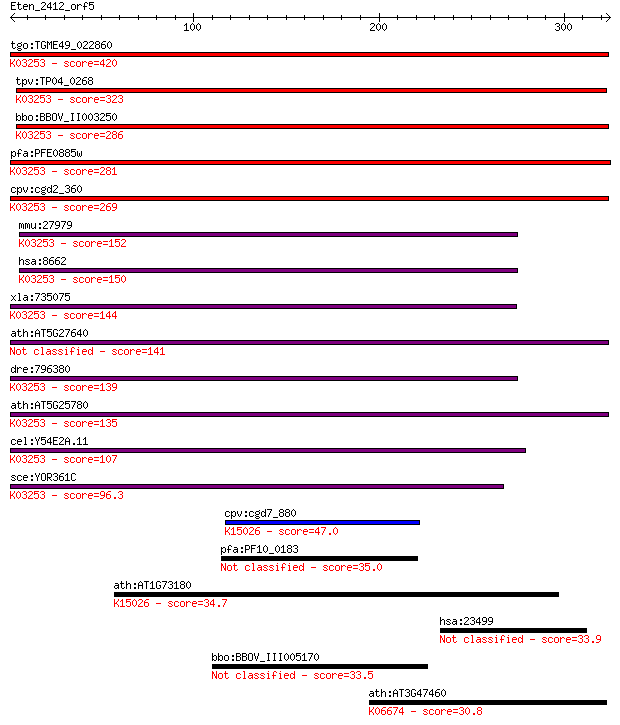
Query= Eten_2412_orf5
Length=324
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_022860 eukaryotic translation initiation factor 3 s... 420 3e-117
tpv:TP04_0268 hypothetical protein; K03253 translation initiat... 323 4e-88
bbo:BBOV_II003250 18.m06273; eukaryotic translation initiation... 286 7e-77
pfa:PFE0885w eukaryotictranslation initiation factor 3 subunit... 281 3e-75
cpv:cgd2_360 prtip-like IF39 eukaryotic translation initiation... 269 8e-72
mmu:27979 Eif3b, AL033316, AL033334, AL033369, AW208965, D5Wsu... 152 2e-36
hsa:8662 EIF3B, EIF3-ETA, EIF3-P110, EIF3-P116, EIF3S9, MGC104... 150 7e-36
xla:735075 eif3b, MGC99017, eIF-3-eta; eukaryotic translation ... 144 7e-34
ath:AT5G27640 TIF3B1; TIF3B1 (TRANSLATION INITIATION FACTOR 3B... 141 4e-33
dre:796380 eif3b, eif3s9, wu:fc17c01; eukaryotic translation i... 139 1e-32
ath:AT5G25780 EIF3B-2 (EUKARYOTIC TRANSLATION INITIATION FACTO... 135 2e-31
cel:Y54E2A.11 eif-3.B; Eukaryotic Initiation Factor family mem... 107 7e-23
sce:YOR361C PRT1, CDC63, DNA26; Prt1p; K03253 translation init... 96.3 2e-19
cpv:cgd7_880 eIF-3A like translation initiation factor that ha... 47.0 1e-04
pfa:PF10_0183 eukaryotic translation initiation factor subunit... 35.0 0.34
ath:AT1G73180 eukaryotic translation initiation factor-related... 34.7 0.46
hsa:23499 MACF1, ABP620, ACF7, FLJ45612, FLJ46776, KIAA0465, K... 33.9 0.95
bbo:BBOV_III005170 17.m07462; hypothetical protein 33.5 1.1
ath:AT3G47460 ATSMC2; ATSMC2; transporter; K06674 structural m... 30.8 7.1
> tgo:TGME49_022860 eukaryotic translation initiation factor 3
subunit 9, putative ; K03253 translation initiation factor
3 subunit B
Length=756
Score = 420 bits (1080), Expect = 3e-117, Method: Compositional matrix adjust.
Identities = 211/325 (64%), Positives = 269/325 (82%), Gaps = 6/325 (1%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
VKGA+MHWQ GDFLCLRT+ FKKTGKKG+KEF+QLEIFRMREKDIPVDN+QLND+AVQL
Sbjct 435 VKGASMHWQSKGDFLCLRTVAFKKTGKKGKKEFTQLEIFRMREKDIPVDNVQLNDVAVQL 494
Query 61 HWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLK 120
HWEEG SKRF LVV +E T+NQ+LRF+RV DA+E KRDTI+ +SFDI YMN+++
Sbjct 495 HWEEGYSKRFALVVHDEQTSNQALRFYRVCDASE----DGKRDTILIYSFDISGYMNYMQ 550
Query 121 WSPFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMP 180
WSPFGSYF+LASL GT++FC LNDQD V+VLH DEH+M NEVRWS CGRYL+T VV+P
Sbjct 551 WSPFGSYFILASLGLDGTLLFCCLNDQDTVQVLHMDEHFMVNEVRWSACGRYLSTAVVLP 610
Query 181 LV--SGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKR 238
++ S ++R G NTG+ IWTFQGKL K +K+ FYQFLWRPHPP+LL ++++++IKK+
Sbjct 611 MLPSSNTASFRLGSNTGYKIWTFQGKLQYKCQKDQFYQFLWRPHPPSLLKKEKIEEIKKK 670
Query 239 MKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQEE 298
MK++SKRYEAEDE+LR E+R AF++QR+EE ++F ++LD L WK+ H Y++W A E
Sbjct 671 MKDYSKRYEAEDEKLRLEQRSAFIRQRKEEMDEFTKVLDGLNQWKVDHELYDDWQLAYET 730
Query 299 FDTWFEWEYKEEIIEEELDVKQEVI 323
FD F+WE KEE+IE+EL+VK+E+I
Sbjct 731 FDAQFDWEDKEEVIEDELEVKEEII 755
> tpv:TP04_0268 hypothetical protein; K03253 translation initiation
factor 3 subunit B
Length=731
Score = 323 bits (829), Expect = 4e-88, Method: Compositional matrix adjust.
Identities = 151/324 (46%), Positives = 222/324 (68%), Gaps = 11/324 (3%)
Query 4 AAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAV-QLHW 62
AMHW GD LCLRT + KKTGKKGRK+F+QLEIFR+RE++IPVD +Q+ D V QLHW
Sbjct 412 VAMHWHSKGDCLCLRTTISKKTGKKGRKQFNQLEIFRLRERNIPVDTIQIQDATVKQLHW 471
Query 63 EEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLKWS 122
EEG SKRF L+V++E T + S+RF++V+D RDT+ T +FDI + +N ++WS
Sbjct 472 EEGGSKRFALIVKDEETRSHSIRFYKVSDVGA------TRDTVWTTTFDITSQLNHMQWS 525
Query 123 PFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPL- 181
P G+YFVL L G++ FC LND DKV+VL++DEH+M N +RWS CGRYLATCV +P+
Sbjct 526 PAGNYFVLGGLGAEGSLFFCALNDNDKVDVLYKDEHFMMNTIRWSSCGRYLATCVNVPMP 585
Query 182 ---VSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKR 238
+ + ++R+ G+ +WTFQG+LL K +KENFY F +RPHP LL +++ IKK
Sbjct 586 QHNATTSDSFRYSAEAGYCLWTFQGRLLFKDRKENFYSFDFRPHPTPLLKRSEIEKIKKN 645
Query 239 MKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQEE 298
+KE++K+Y+ DE+ R + R ++L++R+ + F+Q+ L W Y E+ ++
Sbjct 646 LKEYTKKYDLIDEKERKDYRDSYLQKRQSNLDAFKQLYSSLLQWYTSQDNYLEFKSGWDK 705
Query 299 FDTWFEWEYKEEIIEEELDVKQEV 322
F +WE +++ +E +D+K+EV
Sbjct 706 FLNPLDWEVTDDVYQEVIDIKEEV 729
> bbo:BBOV_II003250 18.m06273; eukaryotic translation initiation
factor 3 subunit; K03253 translation initiation factor 3
subunit B
Length=732
Score = 286 bits (732), Expect = 7e-77, Method: Compositional matrix adjust.
Identities = 141/326 (43%), Positives = 213/326 (65%), Gaps = 12/326 (3%)
Query 4 AAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQ-LHW 62
++MHW G+ CL T +++KTGKKGRK F QL++FR+REK IPVD +++ D V+ L+W
Sbjct 412 SSMHWHPRGECFCLMTTIYRKTGKKGRKNFYQLDLFRLREKYIPVDTIKIEDATVKNLYW 471
Query 63 EEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLKWS 122
EEG +KRF LVV++E + S++F+RV+D E RDT+ ++D+ + MN + WS
Sbjct 472 EEGGNKRFALVVKDEEMSMCSIKFYRVSD------EGTARDTVCVATYDLQSQMNKILWS 525
Query 123 PFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLAT--CVVMP 180
P G+YFVL SLS GT++FC L++ DK+E+L++DEH+M N ++WS CGRYLAT CV MP
Sbjct 526 PTGNYFVLGSLSGEGTLLFCMLSNNDKLEILYKDEHFMMNAMKWSPCGRYLATSVCVPMP 585
Query 181 ---LVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKK 237
LV +R+ GF IWTFQG+ L ++ENFY F +RP PP +L + ++ IKK
Sbjct 586 SQNLVPSTDTFRYSAEAGFCIWTFQGRRLYMARRENFYSFEFRPTPPPMLDKAVIEHIKK 645
Query 238 RMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQE 297
+ E+S+RY+ DE+ R E + + +R+ E F + +L + K ++ ++ R +
Sbjct 646 NLDEYSRRYDIVDEKAREEYEKQYKAKRQVAEEAFIALAAKLMDYLSKQERFNDFKRGYD 705
Query 298 EFDTWFEWEYKEEIIEEELDVKQEVI 323
EF WE +I EE L+VK+E++
Sbjct 706 EFFDESNWEVSYDIFEEVLEVKEEIL 731
> pfa:PFE0885w eukaryotictranslation initiation factor 3 subunit,
putative; K03253 translation initiation factor 3 subunit
B
Length=716
Score = 281 bits (718), Expect = 3e-75, Method: Compositional matrix adjust.
Identities = 143/326 (43%), Positives = 212/326 (65%), Gaps = 8/326 (2%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAV-Q 59
V A++HW GD+LCL+T + KK+GKK +KE++QLEIFRMREK+IPVDN+Q+ + Q
Sbjct 397 VSVASIHWHNKGDYLCLKTTIQKKSGKKIKKEYTQLEIFRMREKNIPVDNVQIEAVKTKQ 456
Query 60 LHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFL 119
HWEE S RF L+++EE T Q +RF+++++ RD + +FDI N MNF+
Sbjct 457 FHWEESNSNRFALILREETTNKQQIRFYKISNKGTA------RDAKWSSTFDINNQMNFM 510
Query 120 KWSPFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVM 179
KWSP G+YF+LASL++ GT+ FC LN+ D+V+V+H+DEH + N + WS CGR+L + V
Sbjct 511 KWSPQGTYFILASLASEGTLYFCCLNNNDEVDVIHKDEHLLVNSISWSRCGRFLVSAVSN 570
Query 180 PLVSGAQAYRFGDN-TGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKR 238
+ YR ++ GF+IWTFQGK L + KK +F QFL+RPHP +L +K DIK
Sbjct 571 NPNAFNTGYREENSEAGFHIWTFQGKCLMRIKKPSFIQFLFRPHPKSLFNDKLKLDIKNN 630
Query 239 MKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQEE 298
+ +SK+++ DE++++ K+ A L +R+ F Q E+ A + +YE+ + E
Sbjct 631 LNNYSKKFDNIDEKMKTAKKNALLTERKNVENSFNQKFQEITALFQSYQEYEQLKKNWET 690
Query 299 FDTWFEWEYKEEIIEEELDVKQEVIC 324
F+ FEWE K +IE L VK+E+
Sbjct 691 FENQFEWEEKTIVIEHVLSVKEEIFA 716
> cpv:cgd2_360 prtip-like IF39 eukaryotic translation initiation
factor 3 ; K03253 translation initiation factor 3 subunit
B
Length=724
Score = 269 bits (688), Expect = 8e-72, Method: Compositional matrix adjust.
Identities = 135/324 (41%), Positives = 217/324 (66%), Gaps = 17/324 (5%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQ- 59
V+ A++HWQ G ++CL+ +V +K GKK +KE++QLEIFR++EK++PVD + + + V+
Sbjct 413 VREASVHWQPKGQYMCLKALVSRKAGKKAKKEYTQLEIFRVKEKNVPVDTVHIEGVTVKC 472
Query 60 LHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFL 119
L WE S RF +VV ++ T +Q+LRF++V N T S+ + + ++ +
Sbjct 473 LSWES--SNRFAIVVVDDVTRSQTLRFYQV----------NATQTDFVCSYYLTSPVDTI 520
Query 120 KWSPFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVM 179
KWSP GSYFVL + G + FC L +++K ++L +DEH+MCN ++W GRY+ T
Sbjct 521 KWSPLGSYFVLGG--SGGNLTFCQLTNENKFDILQKDEHFMCNWIQWDPTGRYVTTAFQS 578
Query 180 PLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRM 239
L GA Y++ TGF +W+FQG+ L + E FYQF+WRPHPP+LL++++ D+I +++
Sbjct 579 KLAEGA--YKYSTETGFIVWSFQGRQLYNSPLETFYQFIWRPHPPSLLSQEKFDEIPRKL 636
Query 240 KEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQEEF 299
K++SK+++AEDE +RSEKR L++R+ ++F IL +++ WK++ P Y+EW A+E
Sbjct 637 KDYSKKFDAEDEAVRSEKRNIVLQKRKVNEDEFNAILQKIQEWKVQQPLYKEWLAAKEVL 696
Query 300 DTWFEWEYKEEIIEEELDVKQEVI 323
+WE +EE+IE EL+ QEVI
Sbjct 697 LNSLQWEEQEEVIEIELETIQEVI 720
> mmu:27979 Eif3b, AL033316, AL033334, AL033369, AW208965, D5Wsu45e,
EIF3-ETA, EIF3-P110, EIF3-P116, Eif3s9, PRT1; eukaryotic
translation initiation factor 3, subunit B; K03253 translation
initiation factor 3 subunit B
Length=803
Score = 152 bits (383), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 93/270 (34%), Positives = 143/270 (52%), Gaps = 28/270 (10%)
Query 6 MHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQLHWEEG 65
+HWQ NGD+LC++ +T K + + EIFRMREK +PVD +++ + + WE
Sbjct 506 LHWQKNGDYLCVKV---DRTPKGTQGVVTNFEIFRMREKQVPVDVVEMKETIIAFAWEPN 562
Query 66 PSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLKWSPFG 125
SK VL + S+ F+ V + G +E + FD N + WSP G
Sbjct 563 GSKFAVLHGE---APRISVSFYHVK--SNGKIE-------LIKMFD-KQQANTIFWSPQG 609
Query 126 SYFVLASL-STYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPLVSG 184
+ VLA L S G + F +D V++ EHYM ++V W GRY+ T V
Sbjct 610 QFVVLAGLRSMNGALAFVDTSD---CTVMNIAEHYMASDVEWDPTGRYVVTSVSW----- 661
Query 185 AQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMKEHSK 244
+ + + +WTFQG+LL+K K+ F Q LWRP PPTLL++ Q+ IKK +K++SK
Sbjct 662 ---WSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQDQIKQIKKDLKKYSK 718
Query 245 RYEAEDERLRSEKRRAFLKQREEESEKFQQ 274
+E +D +S+ + +++R E F+Q
Sbjct 719 IFEQKDRLSQSKASKELVERRRTMMEDFRQ 748
> hsa:8662 EIF3B, EIF3-ETA, EIF3-P110, EIF3-P116, EIF3S9, MGC104664,
MGC131875, PRT1; eukaryotic translation initiation factor
3, subunit B; K03253 translation initiation factor 3 subunit
B
Length=814
Score = 150 bits (378), Expect = 7e-36, Method: Compositional matrix adjust.
Identities = 92/270 (34%), Positives = 144/270 (53%), Gaps = 28/270 (10%)
Query 6 MHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQLHWEEG 65
+HWQ NGD+LC++ +T K + + EIFRMREK +PVD +++ + + WE
Sbjct 517 LHWQKNGDYLCVKV---DRTPKGTQGVVTNFEIFRMREKQVPVDVVEMKETIIAFAWEPN 573
Query 66 PSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLKWSPFG 125
SK VL + S+ F+ V + G +E + FD N + WSP G
Sbjct 574 GSKFAVLHGE---APRISVSFYHVKN--NGKIE-------LIKMFD-KQQANTIFWSPQG 620
Query 126 SYFVLASL-STYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPLVSG 184
+ VLA L S G + F +D V++ EHYM ++V W GRY+ T V
Sbjct 621 QFVVLAGLRSMNGALAFVDTSD---CTVMNIAEHYMASDVEWDPTGRYVVTSVSW----- 672
Query 185 AQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMKEHSK 244
+ + + +WTFQG+LL+K K+ F Q LWRP PPTLL+++Q+ IKK +K++SK
Sbjct 673 ---WSHKVDNAYWLWTFQGRLLQKNNKDRFCQLLWRPRPPTLLSQEQIKQIKKDLKKYSK 729
Query 245 RYEAEDERLRSEKRRAFLKQREEESEKFQQ 274
+E +D +S+ + +++R E F++
Sbjct 730 IFEQKDRLSQSKASKELVERRRTMMEDFRK 759
> xla:735075 eif3b, MGC99017, eIF-3-eta; eukaryotic translation
initiation factor 3, subunit B; K03253 translation initiation
factor 3 subunit B
Length=688
Score = 144 bits (362), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 88/274 (32%), Positives = 143/274 (52%), Gaps = 28/274 (10%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
V +HWQ NGD+LC++ +T K + + EIFRMREK +PVD +++ D +
Sbjct 388 VVDCKLHWQKNGDYLCVKV---DRTPKGTQGMVTNFEIFRMREKQVPVDVVEMKDGIIAF 444
Query 61 HWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLK 120
WE SK VL + S+ F+ V + G +E + +D N +
Sbjct 445 AWEPNGSKFAVLHGE---VPRISVSFYHVKN--NGKIE-------LIKMYD-KQQANTIF 491
Query 121 WSPFGSYFVLASL-STYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVM 179
WSP G + VLA L S G + F +D +++ EHY ++V W GRY+ T V
Sbjct 492 WSPQGQFLVLAGLRSMNGALAFVDTSD---CTIMNIAEHYTASDVEWDPTGRYVITSV-- 546
Query 180 PLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRM 239
+ + + +WTFQG+LL+K K+ F Q LWRP PP+LL++ Q+ IKK +
Sbjct 547 ------SWWSHKVDNAYWMWTFQGRLLQKNNKDRFCQLLWRPRPPSLLSQDQIKQIKKDL 600
Query 240 KEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQ 273
K++SK +E +D +++ + +++R E+++
Sbjct 601 KKYSKIFEQKDRLSQTKASKELIERRRAMMEEYK 634
> ath:AT5G27640 TIF3B1; TIF3B1 (TRANSLATION INITIATION FACTOR
3B1); nucleic acid binding / translation initiation factor
Length=738
Score = 141 bits (355), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 103/333 (30%), Positives = 171/333 (51%), Gaps = 40/333 (12%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQL---NDIA 57
V M+WQ +G++L ++ + KT K +S E+FR++E+DIP++ L+L ND
Sbjct 433 VSDCKMYWQSSGEYLAVKVDRYTKTKKS---TYSGFELFRIKERDIPIEVLELDNKNDKI 489
Query 58 VQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMN 117
+ WE RF ++ ++ + S F A+ +K T+ N
Sbjct 490 IAFAWEPK-GHRFAVIHGDQPRPDVS---FYSMKTAQNTGRVSKLATLKAKQ------AN 539
Query 118 FLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATC 176
L WSP G Y +LA L + G + F ++ D++E + EH+M ++ W GRY+AT
Sbjct 540 ALFWSPTGKYIILAGLKGFNGQLEFFNV---DELETMATAEHFMATDIEWDPTGRYVATA 596
Query 177 VVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIK 236
V + GF IW+F G +L + K++F+Q WRP PP+ LT ++ ++I
Sbjct 597 V-------TSVHEM--ENGFTIWSFNGIMLYRILKDHFFQLAWRPRPPSFLTAEKEEEIA 647
Query 237 KRMKEHSKRYEAEDER---LRSEKRRAFLKQREEESEKFQ---QILDELEAWKMKHPKYE 290
K +K++SK+YEAED+ L SE+ R K +EE EK+ + L E E ++ +
Sbjct 648 KTLKKYSKKYEAEDQDVSLLLSEQDREKRKALKEEWEKWVMQWKSLHEEEKLVRQNLRDG 707
Query 291 EWCRAQEEFDTWFEWEYKEEIIEEELDVKQEVI 323
E +E+ E+E KE E+ +DV +E++
Sbjct 708 EVSDVEED-----EYEAKEVEFEDLIDVTEEIV 735
> dre:796380 eif3b, eif3s9, wu:fc17c01; eukaryotic translation
initiation factor 3, subunit B; K03253 translation initiation
factor 3 subunit B
Length=681
Score = 139 bits (350), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 93/275 (33%), Positives = 146/275 (53%), Gaps = 28/275 (10%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
V +HWQ NGD+LC++ +T K + + EIFRMREK +PVD +++ D +
Sbjct 380 VVDCKLHWQKNGDYLCVKV---DRTPKGTQGVVTNFEIFRMREKQVPVDVVEMKDSIIAF 436
Query 61 HWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMNFLK 120
WE SK F ++ E N S F+ V + G ++ + FD N +
Sbjct 437 AWEPNGSK-FAVLHGESPRINSS--FYHVK--SNGKID-------LIKMFD-KQQANSIF 483
Query 121 WSPFGSYFVLASL-STYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVM 179
WSP G + VLA L S G + F +D +++ EHYM ++V W GR++ T V
Sbjct 484 WSPQGQFLVLAGLRSMNGALAFVDTSD---CTMMNIAEHYMASDVEWDPTGRFVVTSV-- 538
Query 180 PLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRM 239
+ + F +WTFQG+LL+K KE F Q LWRP PP+LLT++Q+ IKK +
Sbjct 539 ------SWWSHKVDNAFWLWTFQGRLLQKNNKERFCQLLWRPRPPSLLTQEQIKLIKKDL 592
Query 240 KEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQ 274
K++SK +E +D +S+ + + +R E++++
Sbjct 593 KKYSKIFEQKDRLSQSKASKELVDKRRTMMEEYRK 627
> ath:AT5G25780 EIF3B-2 (EUKARYOTIC TRANSLATION INITIATION FACTOR
3B-2); nucleic acid binding / protein binding / translation
initiation factor; K03253 translation initiation factor
3 subunit B
Length=714
Score = 135 bits (340), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 100/335 (29%), Positives = 166/335 (49%), Gaps = 44/335 (13%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQL---NDIA 57
V M+WQ +G++L ++ + KT K +S E+FR++E+DIP++ L+L ND
Sbjct 408 VSDCKMYWQSSGEYLAVKVDRYTKTKKS---TYSGFELFRIKERDIPIEVLELDNKNDKI 464
Query 58 VQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIPNYMN 117
+ WE RF ++ ++ + S F A+ +K T+ N
Sbjct 465 IAFAWEP-KGHRFAVIHGDQPRPDVS---FYSMKTAQNTGRVSKLATLKAKQ------AN 514
Query 118 FLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATC 176
L WSP G Y +LA L + G + F ++ D++E + EH+M ++ W GRY+AT
Sbjct 515 ALFWSPTGKYIILAGLKGFNGQLEFFNV---DELETMATAEHFMATDIEWDPTGRYVATS 571
Query 177 VVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIK 236
V V + GF IW+F G ++ + K++F+Q WRP P + LT ++ ++I
Sbjct 572 VTT--VHEME-------NGFTIWSFNGNMVYRILKDHFFQLAWRPRPASFLTAEKEEEIA 622
Query 237 KRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKM--KHPKYEEWCR 294
K ++ +SKRYEAED+ + L E++ EK + + +E + W M K EE
Sbjct 623 KNLRNYSKRYEAEDQDVS-------LLLSEQDREKRRALNEEWQKWVMQWKSLHEEEKLV 675
Query 295 AQEEFDTWF------EWEYKEEIIEEELDVKQEVI 323
Q D E+E KE E+ +DV +E++
Sbjct 676 RQNLRDGEVSDVEEDEYEAKEVEFEDLIDVTEEIV 710
> cel:Y54E2A.11 eif-3.B; Eukaryotic Initiation Factor family member
(eif-3.B); K03253 translation initiation factor 3 subunit
B
Length=725
Score = 107 bits (267), Expect = 7e-23, Method: Compositional matrix adjust.
Identities = 86/299 (28%), Positives = 135/299 (45%), Gaps = 42/299 (14%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFS-------QLEIFRMREKDIPVDNLQL 53
V A M WQ +G L TM FKK + E ++IF + +KD+ + NL L
Sbjct 404 VADAQMFWQKSGKRLAFYTMRFKKKEYRETGEVKYVGGCQYHVDIFEIDKKDVSLMNLPL 463
Query 54 NDIAVQLHWEEGPSKRFVLVVQEEGTTNQSLRFFRVADAAEGNVETNKRDTIMTHSFDIP 113
++ + W+ K VLV T Q + +E N + D
Sbjct 464 SEPFIHFDWDPEGDKFCVLVGNTAKATPQVYK-----------IEANSHAPKLVSKLDAG 512
Query 114 NYMNFLKWSPFGSYF-VLASLSTYGTVMF--CHLNDQDKVEVLHQDEHYMCNEVRWSHCG 170
+ N ++++P G + VLA +S G V F L++ + V+ EH + N+ W G
Sbjct 513 VHFNEVQFAPKGGWLAVLAKVSAGGNVYFIDTSLSEAKRTNVI---EHPLFNKGYWDPTG 569
Query 171 RYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPTLLTEK 230
RY TC + R G + G+ I+TFQG+ L + + QF WRP PP L+E+
Sbjct 570 RYFVTCSTL-------GGRAGADLGYRIFTFQGRELCRKNLDRLAQFKWRPRPPVKLSEQ 622
Query 231 QLDDIKKRMKEHSKRY--EAEDERLRS-----EKRR----AFLKQREEESEKFQQILDE 278
+ +IKK +K+ + ++ + +DE+ R+ EKRR AF R E+ DE
Sbjct 623 KQREIKKNLKKTAAKFIKQDDDEKCRASQEVVEKRRKIMAAFDIIRSRNREQLDATRDE 681
> sce:YOR361C PRT1, CDC63, DNA26; Prt1p; K03253 translation initiation
factor 3 subunit B
Length=763
Score = 96.3 bits (238), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 70/289 (24%), Positives = 129/289 (44%), Gaps = 39/289 (13%)
Query 1 VKGAAMHWQGNGDFLCLRTMVFKKTGKKGRKEFSQLEIFRMREKDIPVDNLQLNDIAVQL 60
V +HWQ +FLC ++ K G+ +FS L+I R+ E+DIPV+ ++L D +
Sbjct 436 VSNVTLHWQNQAEFLCFNV---ERHTKSGKTQFSNLQICRLTERDIPVEKVELKDSVFEF 492
Query 61 HWEEGPSKRFVLVVQEEGTTN-----QSLRFFRVADAAEGNVETNKRDTIMTHSFDIP-N 114
WE ++ + V E N ++RF+ A + + KR +++ +IP
Sbjct 493 GWEPHGNRFVTISVHEVADMNYAIPANTIRFY--APETKEKTDVIKRWSLVK---EIPKT 547
Query 115 YMNFLKWSPFGSYFVLASLSTYGT----VMFC--------HLNDQDKVEVLHQD----EH 158
+ N + WSP G + V+ +L + F ++ND + V +D +
Sbjct 548 FANTVSWSPAGRFVVVGALVGPNMRRSDLQFYDMDYPGEKNINDNNDVSASLKDVAHPTY 607
Query 159 YMCNEVRWSHCGRYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFL 218
+ W GRY+ + + + + G+ I+ G L+++ F F
Sbjct 608 SAATNITWDPSGRYVT--------AWSSSLKHKVEHGYKIFNIAGNLVKEDIIAGFKNFA 659
Query 219 WRPHPPTLLTEKQLDDIKKRMKEHSKRYEAEDE-RLRSEKRRAFLKQRE 266
WRP P ++L+ + ++K ++E S ++E +D + R L QRE
Sbjct 660 WRPRPASILSNAERKKVRKNLREWSAQFEEQDAMEADTAMRDLILHQRE 708
> cpv:cgd7_880 eIF-3A like translation initiation factor that
has a WD40 repeat-containing beta propeller ; K15026 translation
initiation factor 2A
Length=586
Score = 47.0 bits (110), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 43/105 (40%), Gaps = 10/105 (9%)
Query 117 NFLKWSPFGSYFVLASLSTYGTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATC 176
N ++W+P G +F + + D DK +++ Q C + WS CGR+L
Sbjct 392 NTIRWNPSGKWFAYGGFGNLAGDL--DIWDLDKQKLIGQTNASCCITLEWSVCGRFLLAS 449
Query 177 VVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRP 221
P + A R I+ + G LL++ + Y W P
Sbjct 450 TTSPRLRVDNAIR--------IFRYNGDLLKRVNFDTLYSAYWSP 486
> pfa:PF10_0183 eukaryotic translation initiation factor subunit
eIF2A, putative
Length=2405
Score = 35.0 bits (79), Expect = 0.34, Method: Composition-based stats.
Identities = 24/107 (22%), Positives = 38/107 (35%), Gaps = 9/107 (8%)
Query 115 YMNFLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYL 173
Y N +KW PFG+ L G + F + + V ++ + WS+ G
Sbjct 457 YKNTIKWGPFGNMIALGGFGNLAGDINFYYKEVDNSVSIIKKYREACTVLCDWSYDGTLF 516
Query 174 ATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWR 220
T P R F I+T++G L+ Y W+
Sbjct 517 MTASTYP--------RMKVENVFKIYTYEGTLVNSYNFNELYDVKWK 555
> ath:AT1G73180 eukaryotic translation initiation factor-related;
K15026 translation initiation factor 2A
Length=484
Score = 34.7 bits (78), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 58/280 (20%), Positives = 101/280 (36%), Gaps = 66/280 (23%)
Query 57 AVQLHWEEGPSKRFVLVVQEEGTTNQS------LRFFRVADAAEGNVETNKRDTI----- 105
+VQ W G + V+V + TNQS L + + EG V K +
Sbjct 221 SVQFSWNHGSTGLLVVVQSDVDKTNQSYYGETKLHYLTIDGTHEGLVPLRKEGPVHDVQW 280
Query 106 --------MTHSFDIPNYM------------------NFLKWSPFGSYFVLASLSTY-GT 138
+ + F +P + N L+W+P G +A G
Sbjct 281 SFSGSEFAVVYGF-MPACVTIFDKNCKPLMELGEGPYNTLRWNPKGRVLCVAGFGNLPGD 339
Query 139 VMFCHLNDQDKVEVLHQDEHYMCNEVRWSHCGRYLATCVVMPLVSGAQAYRFGDNTGFNI 198
+ F + ++ ++ ++ E + +E WS GRY T P R + G I
Sbjct 340 MAFWDVVNKKQLGS-NKAEWSVTSE--WSPDGRYFLTASTAP--------RRQIDNGMKI 388
Query 199 WTFQGKLLEKTKKENFYQFLWRPHPPTLLTEKQLDDIKKRMK--EHSKRYEAEDERLRSE 256
+ + GK K E YQ W+P P ++ DI + +K E K E + + S
Sbjct 389 FNYDGKRYYKKMFERLYQAEWKPESP-----ERFGDISELIKSVESLKLGEGKSQGQGSA 443
Query 257 KRRAFLKQREEESEKFQQILDELEAWKMKHPKYEEWCRAQ 296
+++ I + A++ H K+ +A+
Sbjct 444 QKKTIAP---------NPIAQKPAAYRPPHAKHAAAVQAE 474
> hsa:23499 MACF1, ABP620, ACF7, FLJ45612, FLJ46776, KIAA0465,
KIAA1251, MACF, OFC4; microtubule-actin crosslinking factor
1
Length=5430
Score = 33.9 bits (76), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 42/82 (51%), Gaps = 5/82 (6%)
Query 233 DDIKKRMKEHSKRYEAEDERLRSEKRRAFLKQREEESEKFQQILDELEAWKMKHPKYEE- 291
+++ K M S + E + ++E +AFL + E+ S K Q++ + L + +P +E
Sbjct 2418 EEVLKSMDAMSSPTKTETVKAQAESNKAFLAELEQNSPKIQKVKEALAGLLVTYPNSQEA 2477
Query 292 --WCRAQEEFDTWFEWEYKEEI 311
W + QEE ++ WE E+
Sbjct 2478 ENWKKIQEELNS--RWERATEV 2497
> bbo:BBOV_III005170 17.m07462; hypothetical protein
Length=734
Score = 33.5 bits (75), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 28/118 (23%), Positives = 44/118 (37%), Gaps = 10/118 (8%)
Query 110 FDIPN-YMNFLKWSPFGSYFVLASLSTY-GTVMFCHLNDQDKVEVLHQDEHYMCNEVRWS 167
F+ P Y N +KW+P G+ L G + F + D + E + Q + WS
Sbjct 419 FEFPKMYRNTIKWNPLGNMVALCGFGNLAGEICFWYRKDNGEYEQIVQFKEPCTVISEWS 478
Query 168 HCGRYLATCVVMPLVSGAQAYRFGDNTGFNIWTFQGKLLEKTKKENFYQFLWRPHPPT 225
+ RY T P R + I++ + LL+ + Y W P T
Sbjct 479 YDARYFITASTFP--------RMKVDNCIKIFSHEADLLDSKTFDECYGVCWIKTPET 528
> ath:AT3G47460 ATSMC2; ATSMC2; transporter; K06674 structural
maintenance of chromosome 2
Length=1171
Score = 30.8 bits (68), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 35/146 (23%), Positives = 62/146 (42%), Gaps = 18/146 (12%)
Query 195 GFNIWTFQGKLLEKTKKENFYQF--LWRPHPPTLLTEKQLDDI--KKRMKEHSKRYEAED 250
G N + GKL + + +N + L +P L+ + ++ + K M+ S EA
Sbjct 111 GKNKYLINGKLAQPNQVQNLFHSVQLNVNNPHFLIMQGRITKVLNMKPMEILSMLEEAAG 170
Query 251 ERLRSEKRRAFLKQREEESEKFQQI--------LDELEAWKMKHPKYEEWCRAQEEFD-- 300
R+ K+ A LK E++ K +I L LE + + +Y +W E D
Sbjct 171 TRMYENKKEAALKTLEKKQTKVDEINKLLEKDILPALEKLRREKSQYMQWANGNAELDRL 230
Query 301 ----TWFEWEYKEEIIEEELDVKQEV 322
FE+ E+I + + V +E+
Sbjct 231 KRFCVAFEYVQAEKIRDNSIHVVEEM 256
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 13394984740
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40