bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2406_orf2
Length=133
Score E
Sequences producing significant alignments: (Bits) Value
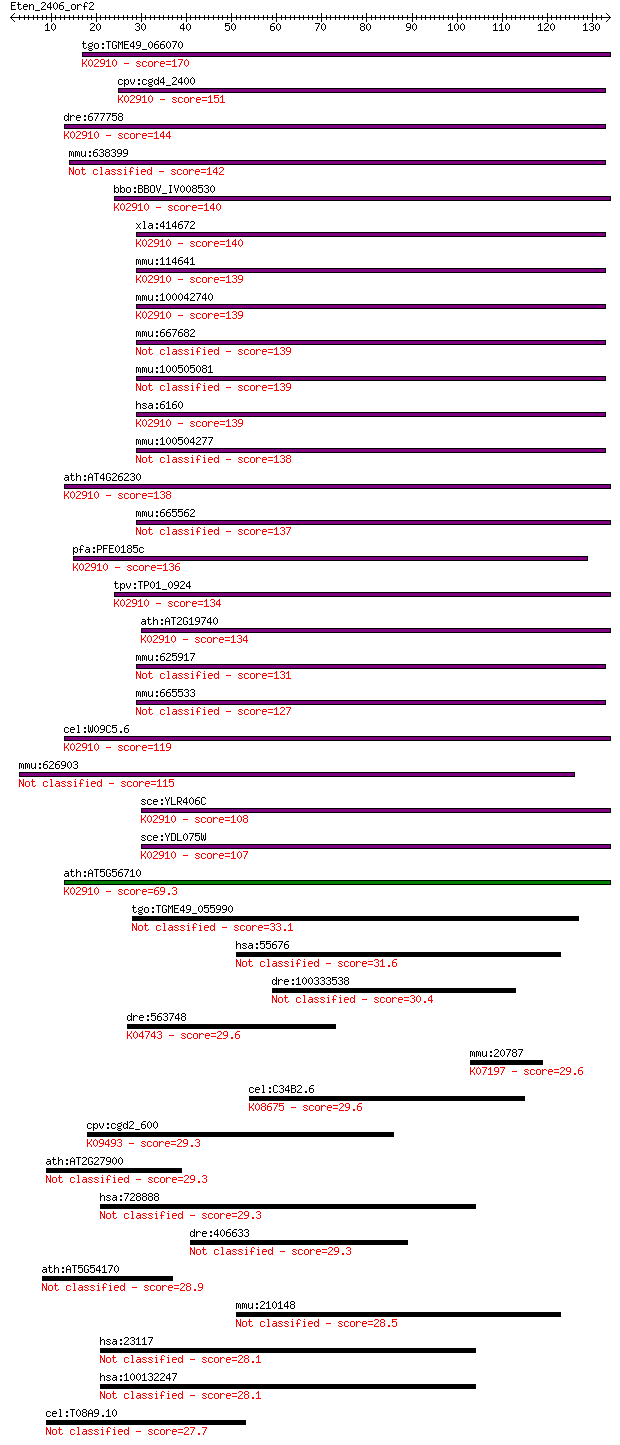
tgo:TGME49_066070 60s ribosomal protein L31, putative ; K02910... 170 1e-42
cpv:cgd4_2400 60S ribosomal protein L31 ; K02910 large subunit... 151 6e-37
dre:677758 MGC136896; zgc:136896; K02910 large subunit ribosom... 144 7e-35
mmu:638399 60S ribosomal protein L31-like 142 3e-34
bbo:BBOV_IV008530 23.m06299; ribosomal protein L31; K02910 lar... 140 1e-33
xla:414672 rpl31, MGC78859; ribosomal protein L31; K02910 larg... 140 1e-33
mmu:114641 Rpl31, MGC106651, MGC107628; ribosomal protein L31;... 139 3e-33
mmu:100042740 Gm10191; predicted pseudogene 10191; K02910 larg... 139 3e-33
mmu:667682 Gm8759, EG667682; predicted gene 8759 139 3e-33
mmu:100505081 60S ribosomal protein L31-like 139 3e-33
hsa:6160 RPL31, MGC88191; ribosomal protein L31; K02910 large ... 139 3e-33
mmu:100504277 ribosomal protein L31 pseudogene 138 4e-33
ath:AT4G26230 60S ribosomal protein L31 (RPL31B); K02910 large... 138 6e-33
mmu:665562 Gm7689, EG665562; predicted pseudogene 7689 137 1e-32
pfa:PFE0185c 60S ribosomal protein L31, putative; K02910 large... 136 1e-32
tpv:TP01_0924 60S ribosomal protein L31; K02910 large subunit ... 134 6e-32
ath:AT2G19740 60S ribosomal protein L31 (RPL31A); K02910 large... 134 1e-31
mmu:625917 Gm6635, EG625917; predicted gene 6635 131 6e-31
mmu:665533 Gm13004, OTTMUSG00000009823; 60S ribosomal protein ... 127 8e-30
cel:W09C5.6 rpl-31; Ribosomal Protein, Large subunit family me... 119 2e-27
mmu:626903 Gm6718, EG626903; predicted gene 6718 115 3e-26
sce:YLR406C RPL31B; Rpl31bp; K02910 large subunit ribosomal pr... 108 5e-24
sce:YDL075W RPL31A, RPL34; Rpl31ap; K02910 large subunit ribos... 107 8e-24
ath:AT5G56710 60S ribosomal protein L31 (RPL31C); K02910 large... 69.3 3e-12
tgo:TGME49_055990 hypothetical protein 33.1 0.26
hsa:55676 SLC30A6, FLJ31101, MGC45055, MST103, MSTP103, ZNT6; ... 31.6 0.61
dre:100333538 adenylyl cyclase 35C-like 30.4 1.6
dre:563748 chondroitin 6-sulfotransferase 7-like; K04743 chond... 29.6 2.7
mmu:20787 Srebf1, ADD-1, ADD1, D630008H06, SREBP-1, SREBP-1a, ... 29.6 2.8
cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependen... 29.6 2.9
cpv:cgd2_600 t-complex protein 1, alpha subunit ; K09493 T-com... 29.3 3.1
ath:AT2G27900 hypothetical protein 29.3 3.2
hsa:728888 nuclear pore complex-interacting protein-like 3-like 29.3 3.2
dre:406633 tesc, fc43f02, wu:fc43f02, zgc:77495; tescalcin 29.3 3.8
ath:AT5G54170 hypothetical protein 28.9 4.0
mmu:210148 Slc30a6, 9530029F08Rik, MGC11963, ZnT6; solute carr... 28.5 5.9
hsa:23117 NPIPL3, FLJ17456, FLJ45371, FLJ54505, FLJ58661, FLJ7... 28.1 7.8
hsa:100132247 DKFZp564D113, FLJ61599, MGC70907; nuclear pore c... 28.1 7.8
cel:T08A9.10 spp-6; SaPosin-like Protein family member (spp-6) 27.7 8.6
> tgo:TGME49_066070 60s ribosomal protein L31, putative ; K02910
large subunit ribosomal protein L31e
Length=120
Score = 170 bits (431), Expect = 1e-42, Method: Compositional matrix adjust.
Identities = 92/117 (78%), Positives = 104/117 (88%), Gaps = 0/117 (0%)
Query 17 SGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKL 76
+ +K KS+QP CRDYTIHLHK++HGIQFKKRAPRA+REIR+FA+K MHTKDVRIDTKL
Sbjct 4 TTRKAQNKSLQPVCRDYTIHLHKLIHGIQFKKRAPRALREIRRFAQKTMHTKDVRIDTKL 63
Query 77 NKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
NK IWS GIRN+PRRVRVR+ARRRN+DED+ EKFYTLVQHVPVASF L EYVNEE
Sbjct 64 NKFIWSGGIRNVPRRVRVRIARRRNDDEDSKEKFYTLVQHVPVASFENLKTEYVNEE 120
> cpv:cgd4_2400 60S ribosomal protein L31 ; K02910 large subunit
ribosomal protein L31e
Length=115
Score = 151 bits (381), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 70/108 (64%), Positives = 87/108 (80%), Gaps = 0/108 (0%)
Query 25 SMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNG 84
SM P RDYTI+L K+VH I FKKRAPRAI+ IR+FA KVM T+DVRID KLNK I+S G
Sbjct 8 SMAPCTRDYTINLSKMVHKISFKKRAPRAIKGIREFAGKVMKTEDVRIDAKLNKFIFSKG 67
Query 85 IRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
+RNLP RVRVR++R+R+E ED+ + YTLVQ++PVA+FAGL E V +
Sbjct 68 VRNLPTRVRVRISRKRSESEDSKDSLYTLVQYIPVATFAGLQTEKVQD 115
> dre:677758 MGC136896; zgc:136896; K02910 large subunit ribosomal
protein L31e
Length=123
Score = 144 bits (363), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 69/122 (56%), Positives = 93/122 (76%), Gaps = 2/122 (1%)
Query 13 MAIKSGKKKATKSM--QPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDV 70
MA K G+KK +S + R+YTI++HK +HGI FK+RAPRA++EIRKFA K M T DV
Sbjct 1 MAPKKGEKKKGRSAINEVVTREYTINIHKRIHGISFKRRAPRALKEIRKFAVKEMGTPDV 60
Query 71 RIDTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYV 130
RIDT+LNK +W+ G+RN+P R+RVR++R+RNEDED+ K YTLV +VPV ++ GL V
Sbjct 61 RIDTRLNKAVWAKGVRNVPYRMRVRLSRKRNEDEDSPNKLYTLVTYVPVTTYKGLQTVNV 120
Query 131 NE 132
+E
Sbjct 121 DE 122
> mmu:638399 60S ribosomal protein L31-like
Length=124
Score = 142 bits (358), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 68/120 (56%), Positives = 91/120 (75%), Gaps = 1/120 (0%)
Query 14 AIKSGKKKATKSM-QPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRI 72
A K G+KK ++ + R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRI
Sbjct 4 AKKGGEKKGRSAINEVVTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRI 63
Query 73 DTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
DT+LNK +W+ GIRN+P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 64 DTRLNKAVWAKGIRNVPYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 123
> bbo:BBOV_IV008530 23.m06299; ribosomal protein L31; K02910 large
subunit ribosomal protein L31e
Length=118
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 71/110 (64%), Positives = 90/110 (81%), Gaps = 0/110 (0%)
Query 24 KSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSN 83
+++QP RDYTIHLHK+VH + FK++AP AI++I++FA K M TKDVRIDT LN++IWSN
Sbjct 9 RALQPITRDYTIHLHKLVHRVSFKRKAPTAIKKIKEFASKAMKTKDVRIDTTLNRYIWSN 68
Query 84 GIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
GIRNLPRRVRVR++R+RN+D+DA E +TLVQHVPV F GL E V E
Sbjct 69 GIRNLPRRVRVRVSRKRNDDDDAKEPMFTLVQHVPVEDFTGLQTEVVASE 118
> xla:414672 rpl31, MGC78859; ribosomal protein L31; K02910 large
subunit ribosomal protein L31e
Length=125
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 64/104 (61%), Positives = 83/104 (79%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HGI FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGIGFKKRAPRALKEIRKFAVKEMRTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV ++ GL V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTNYKGLQTVNVDE 124
> mmu:114641 Rpl31, MGC106651, MGC107628; ribosomal protein L31;
K02910 large subunit ribosomal protein L31e
Length=125
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> mmu:100042740 Gm10191; predicted pseudogene 10191; K02910 large
subunit ribosomal protein L31e
Length=125
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> mmu:667682 Gm8759, EG667682; predicted gene 8759
Length=125
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> mmu:100505081 60S ribosomal protein L31-like
Length=125
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> hsa:6160 RPL31, MGC88191; ribosomal protein L31; K02910 large
subunit ribosomal protein L31e
Length=125
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVGFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> mmu:100504277 ribosomal protein L31 pseudogene
Length=125
Score = 138 bits (348), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 63/104 (60%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKKRAPRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 21 VTREYTINIHKRIHGVDFKKRAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYRIRVRLSRKRNEDEDSPNKLYTLVIYVPVTTFKNLQTVNVDE 124
> ath:AT4G26230 60S ribosomal protein L31 (RPL31B); K02910 large
subunit ribosomal protein L31e
Length=119
Score = 138 bits (347), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 67/123 (54%), Positives = 90/123 (73%), Gaps = 7/123 (5%)
Query 13 MAIKSGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRI 72
M+ K G+K+ + R+YTI+LH+ +H FKK+AP+AI+EIRKFAEK M TKDVR+
Sbjct 1 MSEKKGRKE-----EVVTREYTINLHRRLHSCTFKKKAPKAIKEIRKFAEKEMGTKDVRV 55
Query 73 DTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQ--HVPVASFAGLGPEYV 130
D KLNK IWS GIR PRR+RVR+AR+RN+DEDA E+F++LV +P +GLG + +
Sbjct 56 DVKLNKQIWSKGIRGPPRRIRVRVARKRNDDEDAKEEFFSLVTVAEIPAEGLSGLGTKII 115
Query 131 NEE 133
EE
Sbjct 116 EEE 118
> mmu:665562 Gm7689, EG665562; predicted pseudogene 7689
Length=158
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 62/105 (59%), Positives = 81/105 (77%), Gaps = 0/105 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
R+YTI++HK +HG+ FKK APRA++EIRKFA K M T DVRIDT+LNK +W+ GIRN+
Sbjct 54 VTREYTINIHKRIHGVGFKKHAPRALKEIRKFAMKEMGTPDVRIDTRLNKAVWAKGIRNV 113
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
P R+RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 114 PYRIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDEN 158
> pfa:PFE0185c 60S ribosomal protein L31, putative; K02910 large
subunit ribosomal protein L31e
Length=120
Score = 136 bits (343), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 63/114 (55%), Positives = 86/114 (75%), Gaps = 0/114 (0%)
Query 15 IKSGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDT 74
+K KK K+++P + TI+L K+ H + +K++APRAI+EIR A K+MHTKDVR+D
Sbjct 2 VKGTVKKQKKTLKPVTKFITINLSKLTHKVCYKRKAPRAIKEIRSIAGKLMHTKDVRLDV 61
Query 75 KLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPE 128
KLNK IWS G+RN P+RVRV++ R+RNEDED+ EK YT+V+HV V S+ GL E
Sbjct 62 KLNKFIWSKGVRNPPKRVRVKLERKRNEDEDSKEKMYTIVEHVMVDSYKGLVNE 115
> tpv:TP01_0924 60S ribosomal protein L31; K02910 large subunit
ribosomal protein L31e
Length=118
Score = 134 bits (338), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 68/110 (61%), Positives = 89/110 (80%), Gaps = 0/110 (0%)
Query 24 KSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSN 83
K++ P RDYTIHLHK+VH + FK++AP A+++I+ FA + M TKDVR+DT+LN+ +WSN
Sbjct 9 KTLAPLTRDYTIHLHKMVHRVTFKRKAPTAVKKIKAFAARAMKTKDVRLDTRLNEFLWSN 68
Query 84 GIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
GI+NLPRRVRVR++RRRN+DEDA E YTLVQH+PV F+GL E V E
Sbjct 69 GIKNLPRRVRVRVSRRRNDDEDAKEPMYTLVQHIPVDDFSGLQTEVVANE 118
> ath:AT2G19740 60S ribosomal protein L31 (RPL31A); K02910 large
subunit ribosomal protein L31e
Length=119
Score = 134 bits (336), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 62/106 (58%), Positives = 80/106 (75%), Gaps = 2/106 (1%)
Query 30 CRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLP 89
R+YTI+LH+ +H FKK+AP AI+EIRKFA K M TKDVR+D KLNK IWS GIR P
Sbjct 13 TREYTINLHRRLHSCTFKKKAPNAIKEIRKFALKAMGTKDVRVDVKLNKQIWSKGIRGPP 72
Query 90 RRVRVRMARRRNEDEDAAEKFYTLVQ--HVPVASFAGLGPEYVNEE 133
RR+RVR+AR+RN+DEDA E+F++LV +P +GLG + + EE
Sbjct 73 RRIRVRVARKRNDDEDAKEEFFSLVTVAEIPAEGLSGLGTKVIEEE 118
> mmu:625917 Gm6635, EG625917; predicted gene 6635
Length=125
Score = 131 bits (329), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 59/104 (56%), Positives = 80/104 (76%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
++YTI++HK +HG+ F+KRAPRA++EIRKFA K M T DV IDT+LNK +W+ GIRN+
Sbjct 21 VTQEYTINIHKRIHGVGFQKRAPRALKEIRKFAMKEMGTPDVHIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P R RVR++R+RNEDED+ +K YTLV + PV +F L V+E
Sbjct 81 PYRSRVRLSRKRNEDEDSPDKLYTLVTYAPVTTFKNLQTVNVDE 124
> mmu:665533 Gm13004, OTTMUSG00000009823; 60S ribosomal protein
L31 pseudogene
Length=125
Score = 127 bits (320), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 58/104 (55%), Positives = 78/104 (75%), Gaps = 0/104 (0%)
Query 29 ACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
++YTI++HK +HG+ FKK AP A++EIRKFA K M T DV IDT+LNK +W+ GIRN+
Sbjct 21 VTQEYTINIHKRIHGVGFKKSAPWALKEIRKFAMKEMGTPDVLIDTRLNKAVWAKGIRNV 80
Query 89 PRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
P +RVR++R+RNEDED+ K YTLV +VPV +F L V+E
Sbjct 81 PYPIRVRLSRKRNEDEDSPNKLYTLVTYVPVTTFKNLQTVNVDE 124
> cel:W09C5.6 rpl-31; Ribosomal Protein, Large subunit family
member (rpl-31); K02910 large subunit ribosomal protein L31e
Length=122
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 69/121 (57%), Positives = 87/121 (71%), Gaps = 0/121 (0%)
Query 13 MAIKSGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRI 72
MA K+ KK + + R+YTIH+H + GI KKRAPRAI EI+KFA+ M T DVR+
Sbjct 1 MAPKNEKKSRSTINEVVTREYTIHIHARIRGIGSKKRAPRAIDEIKKFAKIQMKTNDVRV 60
Query 73 DTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNE 132
DTKLNK IWS GI+N+P RVRVR++RRRNEDED+A+K YTL +VP +F GL V+
Sbjct 61 DTKLNKFIWSKGIKNVPYRVRVRLSRRRNEDEDSAQKLYTLCTYVPCTNFHGLTNVNVDS 120
Query 133 E 133
E
Sbjct 121 E 121
> mmu:626903 Gm6718, EG626903; predicted gene 6718
Length=130
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 63/125 (50%), Positives = 84/125 (67%), Gaps = 4/125 (3%)
Query 3 SYLQFQQLAKMAIKSGKKKATKSM--QPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKF 60
S L +LA A K G++K +S + R+YTI++HK +HG+ FKK PRA+ EI KF
Sbjct 8 SNLGLVELAP-AKKGGERKKGRSAINKVVTREYTINIHKRIHGVGFKKHVPRALTEILKF 66
Query 61 AEKVMHTKDVRIDTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQHVPVA 120
A K M T DVRIDT+LNK IW GI N+P R RVR++R+ NEDED+++K YT +PV
Sbjct 67 AMKEMGTPDVRIDTRLNKTIWVKGIMNVPHRSRVRLSRKCNEDEDSSDKLYT-GNLLPVT 125
Query 121 SFAGL 125
+F L
Sbjct 126 TFKNL 130
> sce:YLR406C RPL31B; Rpl31bp; K02910 large subunit ribosomal
protein L31e
Length=113
Score = 108 bits (269), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 76/104 (73%), Gaps = 0/104 (0%)
Query 30 CRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLP 89
R+YTI+LHK +HG+ FKKRAPRA++EI+KFA+ M T+DVR+ +LN+ IW G++ +
Sbjct 9 TREYTINLHKRLHGVSFKKRAPRAVKEIKKFAKLHMGTEDVRLAPELNQAIWKRGVKGVE 68
Query 90 RRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
R+R+R++R+RNE+EDA ++ V+ V VAS GL V E+
Sbjct 69 YRLRLRISRKRNEEEDAKNPLFSYVEPVLVASAKGLQTVVVEED 112
> sce:YDL075W RPL31A, RPL34; Rpl31ap; K02910 large subunit ribosomal
protein L31e
Length=113
Score = 107 bits (268), Expect = 8e-24, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 75/104 (72%), Gaps = 0/104 (0%)
Query 30 CRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLP 89
R+YTI+LHK +HG+ FKKRAPRA++EI+KFA+ M T DVR+ +LN+ IW G++ +
Sbjct 9 TREYTINLHKRLHGVSFKKRAPRAVKEIKKFAKLHMGTDDVRLAPELNQAIWKRGVKGVE 68
Query 90 RRVRVRMARRRNEDEDAAEKFYTLVQHVPVASFAGLGPEYVNEE 133
R+R+R++R+RNE+EDA ++ V+ V VAS GL V E+
Sbjct 69 YRLRLRISRKRNEEEDAKNPLFSYVEPVLVASAKGLQTVVVEED 112
> ath:AT5G56710 60S ribosomal protein L31 (RPL31C); K02910 large
subunit ribosomal protein L31e
Length=85
Score = 69.3 bits (168), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 43/123 (34%), Positives = 62/123 (50%), Gaps = 41/123 (33%)
Query 13 MAIKSGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRI 72
M+ K G+K+ + R+YTI+LH+ +H FKK+AP+AI+EIRKFAEK M TKDVR+
Sbjct 1 MSEKKGRKE-----EVITREYTINLHRRLHKCTFKKKAPKAIKEIRKFAEKAMGTKDVRV 55
Query 73 DTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYTLVQ--HVPVASFAGLGPEYV 130
D E+F++LV +P +GLG + +
Sbjct 56 D----------------------------------EEFFSLVTVAEIPAEGLSGLGTKVI 81
Query 131 NEE 133
EE
Sbjct 82 EEE 84
> tgo:TGME49_055990 hypothetical protein
Length=153
Score = 33.1 bits (74), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 47/106 (44%), Gaps = 14/106 (13%)
Query 28 PACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRN 87
P R + + +V + +K+ A++++RK +K++ D I+TK H W
Sbjct 11 PQFRSAVLQVESLVVKPRLRKQDIAALQQLRKKVQKLLKDLDQAINTKHEFHFW------ 64
Query 88 LPRRVRVR-MARRRNEDEDA--AEKFYTLVQHV----PVASFAGLG 126
P R+ V RR E + A L+QH+ P S GLG
Sbjct 65 -PVRLTVEACCFRRTETRQSGKARTVVPLIQHIPRPAPATSNIGLG 109
> hsa:55676 SLC30A6, FLJ31101, MGC45055, MST103, MSTP103, ZNT6;
solute carrier family 30 (zinc transporter), member 6; K14693
solute carrier family 30 (zinc transporter), member 6
Length=501
Score = 31.6 bits (70), Expect = 0.61, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 5/75 (6%)
Query 51 PRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLPRRVRVRMARRRNED---EDAA 107
P I ++ K +V T D ++ + N+H W+ G +L V VR+ R NE
Sbjct 300 PHVIGQLDKLIREV-STLDGVLEVR-NEHFWTLGFGSLAGSVHVRIRRDANEQMVLAHVT 357
Query 108 EKFYTLVQHVPVASF 122
+ YTLV + V F
Sbjct 358 NRLYTLVSTLTVQIF 372
> dre:100333538 adenylyl cyclase 35C-like
Length=597
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 59 KFAEKVMHTKDVRIDTKLNKHIWSNGIRNLPRRVRVRMARRRNEDEDAAEKFYT 112
K + +MH KD+ ++ L + + + +PRRV + ++ ++D D + K Y+
Sbjct 337 KVGQAIMHGKDLEVEKALKERMIHS---VMPRRVADELMKQGDDDNDNSVKRYS 387
> dre:563748 chondroitin 6-sulfotransferase 7-like; K04743 chondroitin
6-sulfotransferase 7 [EC:2.8.2.17]
Length=416
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 18/48 (37%), Positives = 25/48 (52%), Gaps = 2/48 (4%)
Query 27 QPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEK--VMHTKDVRI 72
+P C Y H +V G K P+ +RE+ K +K VM KDVR+
Sbjct 169 EPLCNAYKKHEIGLVQGDVCSKCQPKDLRELEKECKKYPVMVIKDVRV 216
> mmu:20787 Srebf1, ADD-1, ADD1, D630008H06, SREBP-1, SREBP-1a,
SREBP-1c, SREBP1, SREBP1c, bHLHd1; sterol regulatory element
binding transcription factor 1; K07197 sterol regulatory
element-binding transcription factor 1
Length=1134
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 10/16 (62%), Positives = 14/16 (87%), Gaps = 0/16 (0%)
Query 103 DEDAAEKFYTLVQHVP 118
DE+AAE+ Y LV+H+P
Sbjct 877 DEEAAERLYPLVEHIP 892
> cel:C34B2.6 hypothetical protein; K08675 Lon-like ATP-dependent
protease [EC:3.4.21.-]
Length=971
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 17/68 (25%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query 54 IREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLP-------RRVRVRMARRRNEDEDA 106
+R+ A + + +D ++ + + +G+RNL R+ +++A ++NEDE+
Sbjct 668 LRKDTSLATEQLKIEDSALEELIKHYCRESGVRNLQQHIERIFRKAALQIAEQQNEDEEP 727
Query 107 AEKFYTLV 114
AEK T +
Sbjct 728 AEKATTAI 735
> cpv:cgd2_600 t-complex protein 1, alpha subunit ; K09493 T-complex
protein 1 subunit alpha
Length=567
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 18/69 (26%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query 18 GKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTKDVRI-DTKL 76
G + K+++ AC D+ + +++ GI+ + P+ + IR ++V+H + +I T
Sbjct 229 GMPTSVKNVKIACIDFPLKQYRMQMGIRVELEDPKELARIRLEEKEVIHKRIEKILATGC 288
Query 77 NKHIWSNGI 85
N + S GI
Sbjct 289 NVVLTSGGI 297
> ath:AT2G27900 hypothetical protein
Length=1124
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 9 QLAKMAIKSGKKKATKSMQPACRDYTIHLH 38
++A + K+G++ T SM A RD +H H
Sbjct 208 KIANVICKNGRRNLTSSMNEASRDLIVHTH 237
> hsa:728888 nuclear pore complex-interacting protein-like 3-like
Length=1122
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 23/89 (25%), Positives = 39/89 (43%), Gaps = 6/89 (6%)
Query 21 KATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTK--DVRIDTKLNK 78
++ + Q R +H K V + K + + +R+ E + K ++ TK+N+
Sbjct 109 RSNRRRQEGLRSICMHTKKRVSSFRGNKIGLKDVITLRRHVETKVRAKIRKRKVTTKINR 168
Query 79 HIWSNGIRNLPRR----VRVRMARRRNED 103
H NG R R+ R + RRR ED
Sbjct 169 HDKINGKRKTARKQKMFQRAQELRRRAED 197
> dre:406633 tesc, fc43f02, wu:fc43f02, zgc:77495; tescalcin
Length=218
Score = 29.3 bits (64), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 13/48 (27%), Positives = 23/48 (47%), Gaps = 0/48 (0%)
Query 41 VHGIQFKKRAPRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNL 88
V + + +P E F + + KD+ I+TK+N H W+ R +
Sbjct 167 VASVTVGRMSPDEFYEGITFEQFMQILKDIEIETKMNVHFWNLDTRTM 214
> ath:AT5G54170 hypothetical protein
Length=449
Score = 28.9 bits (63), Expect = 4.0, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 8 QQLAKMAIKSGKKKATKSMQPACRDYTIH 36
+++AK+ +K G A K M+P R Y H
Sbjct 312 REIAKLGVKRGMWGAVKKMEPGLRAYQTH 340
> mmu:210148 Slc30a6, 9530029F08Rik, MGC11963, ZnT6; solute carrier
family 30 (zinc transporter), member 6; K14693 solute
carrier family 30 (zinc transporter), member 6
Length=460
Score = 28.5 bits (62), Expect = 5.9, Method: Composition-based stats.
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 5/75 (6%)
Query 51 PRAIREIRKFAEKVMHTKDVRIDTKLNKHIWSNGIRNLPRRVRVRMARRRNED---EDAA 107
P I ++ K +V T D ++ + N+H W+ G +L V VR+ R NE +
Sbjct 260 PHVIGQLDKLIREV-STLDGVLEVR-NEHFWTLGFGSLAGSVHVRIRRDANEQMVLAHVS 317
Query 108 EKFYTLVQHVPVASF 122
+ TLV + V F
Sbjct 318 NRLCTLVSTLTVQIF 332
> hsa:23117 NPIPL3, FLJ17456, FLJ45371, FLJ54505, FLJ58661, FLJ79358,
FLJ98670, KIAA0220, KIAA0220L, MGC105000, MGC70907;
nuclear pore complex interacting protein-like 3
Length=1133
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 23/89 (25%), Positives = 38/89 (42%), Gaps = 6/89 (6%)
Query 21 KATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTK--DVRIDTKLNK 78
++ + Q R +H K V + K + + +R+ E + K ++ TK+N
Sbjct 109 RSNRRRQEGLRSICMHTKKRVSSFRGNKIGLKDVITLRRHVETKVRAKIRKRKVTTKINH 168
Query 79 HIWSNGIRNLPRR----VRVRMARRRNED 103
H NG R R+ R + RRR ED
Sbjct 169 HDKINGKRKTARKQKMFQRAQELRRRAED 197
> hsa:100132247 DKFZp564D113, FLJ61599, MGC70907; nuclear pore
complex interacting protein related gene
Length=1133
Score = 28.1 bits (61), Expect = 7.8, Method: Composition-based stats.
Identities = 23/89 (25%), Positives = 38/89 (42%), Gaps = 6/89 (6%)
Query 21 KATKSMQPACRDYTIHLHKIVHGIQFKKRAPRAIREIRKFAEKVMHTK--DVRIDTKLNK 78
++ + Q R +H K V + K + + +R+ E + K ++ TK+N
Sbjct 109 RSNRRRQEGLRSICMHTKKRVSSFRGNKIGLKDVITLRRHVETKVRAKIRKRKVTTKINH 168
Query 79 HIWSNGIRNLPRR----VRVRMARRRNED 103
H NG R R+ R + RRR ED
Sbjct 169 HDKINGKRKTARKQKMFQRAQELRRRAED 197
> cel:T08A9.10 spp-6; SaPosin-like Protein family member (spp-6)
Length=90
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 3/44 (6%)
Query 9 QLAKMAIKSGKKKATKSMQPACRDYTIHLHKIVHGIQFKKRAPR 52
++ K+A+KS A K +D+ K H IQF APR
Sbjct 16 EMCKLAVKSADGDADKDTNDIKKDFDAKCKKAFHSIQF---APR 56
Lambda K H
0.322 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2165519004
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40