bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2342_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
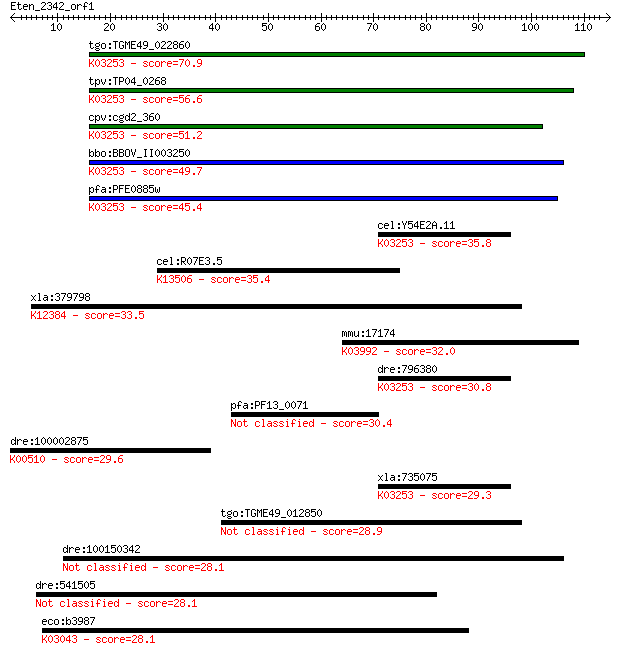
tgo:TGME49_022860 eukaryotic translation initiation factor 3 s... 70.9 1e-12
tpv:TP04_0268 hypothetical protein; K03253 translation initiat... 56.6 2e-08
cpv:cgd2_360 prtip-like IF39 eukaryotic translation initiation... 51.2 8e-07
bbo:BBOV_II003250 18.m06273; eukaryotic translation initiation... 49.7 2e-06
pfa:PFE0885w eukaryotictranslation initiation factor 3 subunit... 45.4 4e-05
cel:Y54E2A.11 eif-3.B; Eukaryotic Initiation Factor family mem... 35.8 0.037
cel:R07E3.5 acl-5; ACyLtransferase-like family member (acl-5);... 35.4 0.051
xla:379798 scarb2, MGC53174, cd36l2; scavenger receptor class ... 33.5 0.16
mmu:17174 Masp1, AW048060, CCPII, Crarf, Masp1/3; mannan-bindi... 32.0 0.44
dre:796380 eif3b, eif3s9, wu:fc17c01; eukaryotic translation i... 30.8 1.00
pfa:PF13_0071 probable protein, unknown function 30.4 1.5
dre:100002875 MGC162585, im:6909100; si:dkey-44g23.7 (EC:1.14.... 29.6 2.5
xla:735075 eif3b, MGC99017, eIF-3-eta; eukaryotic translation ... 29.3 3.1
tgo:TGME49_012850 hypothetical protein 28.9 4.5
dre:100150342 si:dkey-177p2.10; si:dkey-177p2.16 28.1 6.7
dre:541505 cep68, si:ch211-101n13.4, si:zc101n13.4, zgc:112945... 28.1 7.2
eco:b3987 rpoB, ECK3978, ftsR, groN, JW3950, mbrD?, nitB, rif,... 28.1 7.5
> tgo:TGME49_022860 eukaryotic translation initiation factor 3
subunit 9, putative ; K03253 translation initiation factor
3 subunit B
Length=756
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 47/102 (46%), Positives = 66/102 (64%), Gaps = 14/102 (13%)
Query 16 MVKVTKEDLEKLEIKEEEQEELLKYISDDSD--------EDEEALTKKLEAFEPLLQMDD 67
MV++ K+DL E+++LL+++S+DSD + EE L +KL EPLLQMD
Sbjct 1 MVRIDKDDLPA------EEQDLLEFLSEDSDEGEDDDSEQGEEKLKEKLSQMEPLLQMDY 54
Query 68 RFPDTVIMVGVPLVGADRLEKLTAVLRKKIADELSRKGGEDV 109
FP T+++VGVP VG D+ EKL VL KK+ +EL +KG E V
Sbjct 55 SFPSTIVLVGVPKVGKDKHEKLRLVLDKKMGEELLKKGAESV 96
> tpv:TP04_0268 hypothetical protein; K03253 translation initiation
factor 3 subunit B
Length=731
Score = 56.6 bits (135), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 32/92 (34%), Positives = 57/92 (61%), Gaps = 4/92 (4%)
Query 16 MVKVTKEDLEKLEIKEEEQEELLKYISDDSDEDEEALTKKLEAFEPLLQMDDRFPDTVIM 75
MVKV+ DL + EE + LL ++SDD +E +++ + +P + +D F T+++
Sbjct 1 MVKVSPSDLSQ----EELEIGLLDWVSDDDEEFDKSQLDRFYELDPPVTLDKSFKRTLMV 56
Query 76 VGVPLVGADRLEKLTAVLRKKIADELSRKGGE 107
+G+P+VG ++ ++L V+RK I EL +KG E
Sbjct 57 LGLPVVGPEKHDRLRDVVRKTITKELEKKGCE 88
> cpv:cgd2_360 prtip-like IF39 eukaryotic translation initiation
factor 3 ; K03253 translation initiation factor 3 subunit
B
Length=724
Score = 51.2 bits (121), Expect = 8e-07, Method: Composition-based stats.
Identities = 30/89 (33%), Positives = 48/89 (53%), Gaps = 10/89 (11%)
Query 16 MVKVTKEDLEKLEIKEEEQEELLKYISD---DSDEDEEALTKKLEAFEPLLQMDDRFPDT 72
M KVT+E+L E +LL+Y+SD D +E EE ++K+ E L + D F +
Sbjct 1 MFKVTREELG-------EDIDLLEYLSDSTYDGEETEEYISKRCNTIEEPLTLSDSFSKS 53
Query 73 VIMVGVPLVGADRLEKLTAVLRKKIADEL 101
++ G+P +G+D+ E+ LR I L
Sbjct 54 FVIFGLPCIGSDKYERFLKALRTIINQTL 82
> bbo:BBOV_II003250 18.m06273; eukaryotic translation initiation
factor 3 subunit; K03253 translation initiation factor 3
subunit B
Length=732
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 53/90 (58%), Gaps = 4/90 (4%)
Query 16 MVKVTKEDLEKLEIKEEEQEELLKYISDDSDEDEEALTKKLEAFEPLLQMDDRFPDTVIM 75
MV + + DL E+ + L++++SD D +E ++L E +++ FP+T+I+
Sbjct 1 MVVIRESDLSA----EDLADGLMEWMSDHDDSKDEEEMERLALMEHPIELSTIFPNTIIV 56
Query 76 VGVPLVGADRLEKLTAVLRKKIADELSRKG 105
G+PLV ++ ++L +LRKK+ + +KG
Sbjct 57 TGLPLVTQEKYDRLMEILRKKLLKDFEKKG 86
> pfa:PFE0885w eukaryotictranslation initiation factor 3 subunit,
putative; K03253 translation initiation factor 3 subunit
B
Length=716
Score = 45.4 bits (106), Expect = 4e-05, Method: Composition-based stats.
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 9/90 (10%)
Query 16 MVKVTKEDLEKLEIKEEEQEELLKYISDDSDE-DEEALTKKLEAFEPLLQMDDRFPDTVI 74
MV+V+ +L +EL+ ++S+DSD DE L KK E L+ ++ FP V
Sbjct 1 MVRVSDNELS--------DKELVDFLSEDSDNGDENLLNKKYSDKEALITLEVDFPKIVT 52
Query 75 MVGVPLVGADRLEKLTAVLRKKIADELSRK 104
++G+P V +++ +L VL+K LS K
Sbjct 53 ILGIPKVESEKHGRLAEVLKKLFIRHLSAK 82
> cel:Y54E2A.11 eif-3.B; Eukaryotic Initiation Factor family member
(eif-3.B); K03253 translation initiation factor 3 subunit
B
Length=725
Score = 35.8 bits (81), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 71 DTVIMVGVPLVGADRLEKLTAVLRK 95
+ V + G+P+VGADRL KL +VL+K
Sbjct 46 NCVFIAGIPVVGADRLGKLQSVLKK 70
> cel:R07E3.5 acl-5; ACyLtransferase-like family member (acl-5);
K13506 glycerol-3-phosphate O-acyltransferase 3/4 [EC:2.3.1.15]
Length=512
Score = 35.4 bits (80), Expect = 0.051, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 28/46 (60%), Gaps = 2/46 (4%)
Query 29 IKEEEQEELLKYISDDSDEDEEALTKKLEAFEPLLQMDDRFPDTVI 74
I EEE++ELLK I D +DEE + +K+ + +P ++ D D I
Sbjct 466 ISEEERDELLKQI--DEQDDEEEMIRKISSLKPKFKIGDHDADEHI 509
> xla:379798 scarb2, MGC53174, cd36l2; scavenger receptor class
B, member 2; K12384 lysosome membrane protein 2
Length=484
Score = 33.5 bits (75), Expect = 0.16, Method: Composition-based stats.
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 5/96 (5%)
Query 5 FHVFGAENPEKMVKVTK---EDLEKLEIKEEEQEELLKYISDDSDEDEEALTKKLEAFEP 61
F+ F NP +++ K ++ +E Q+E + + ++++ A+T K FEP
Sbjct 64 FYFFNVNNPLEILNGEKPFVTEIGPYTYREYRQKENITFSVNETEV--SAVTPKTYVFEP 121
Query 62 LLQMDDRFPDTVIMVGVPLVGADRLEKLTAVLRKKI 97
+ + D D + V +PLV + K + +LR I
Sbjct 122 EMSVGDPKVDLIRTVNIPLVTVMEMTKDSRILRPLI 157
> mmu:17174 Masp1, AW048060, CCPII, Crarf, Masp1/3; mannan-binding
lectin serine peptidase 1 (EC:3.4.21.-); K03992 mannan-binding
lectin serine protease 1 [EC:3.4.21.-]
Length=704
Score = 32.0 bits (71), Expect = 0.44, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 29/49 (59%), Gaps = 4/49 (8%)
Query 64 QMDDRFPDTVIMVGVPLVGADRLEKLTAVLRKKIADEL----SRKGGED 108
Q RFP+ ++ + +P+V +D ++ L+KK+ ++ ++GG+D
Sbjct 597 QFLQRFPENLMEIEIPIVNSDTCQEAYTPLKKKVTKDMICAGEKEGGKD 645
> dre:796380 eif3b, eif3s9, wu:fc17c01; eukaryotic translation
initiation factor 3, subunit B; K03253 translation initiation
factor 3 subunit B
Length=681
Score = 30.8 bits (68), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 1/26 (3%)
Query 71 DTVIMV-GVPLVGADRLEKLTAVLRK 95
D+VI+V VP VG DRLEKL V+ K
Sbjct 52 DSVIVVDNVPQVGPDRLEKLKNVIHK 77
> pfa:PF13_0071 probable protein, unknown function
Length=562
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 43 DDSDEDEEALTKKLEAFEPLLQMDDRFP 70
+D D++E A TK L A + LL +DD FP
Sbjct 379 EDEDDEEYATTKFLRAHDILLSVDDHFP 406
> dre:100002875 MGC162585, im:6909100; si:dkey-44g23.7 (EC:1.14.99.3);
K00510 heme oxygenase [EC:1.14.99.3]
Length=310
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 12/38 (31%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 1 GLAFFHVFGAENPEKMVKVTKEDLEKLEIKEEEQEELL 38
GL F+H G NP ++ + + +LE+ E + +LL
Sbjct 178 GLNFYHFEGIHNPTAFKRLYRSRMNELEVDAETKAKLL 215
> xla:735075 eif3b, MGC99017, eIF-3-eta; eukaryotic translation
initiation factor 3, subunit B; K03253 translation initiation
factor 3 subunit B
Length=688
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 16/26 (61%), Positives = 19/26 (73%), Gaps = 1/26 (3%)
Query 71 DTVIMV-GVPLVGADRLEKLTAVLRK 95
D+VI+V VP VG DRLEKL V+ K
Sbjct 60 DSVIVVDNVPQVGPDRLEKLKNVIVK 85
> tgo:TGME49_012850 hypothetical protein
Length=414
Score = 28.9 bits (63), Expect = 4.5, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 30/58 (51%), Gaps = 1/58 (1%)
Query 41 ISDDSDEDEEALTKKLEAFEPLLQMDDRFPD-TVIMVGVPLVGADRLEKLTAVLRKKI 97
+SD + E LEA EP+L++D R + V++ G+D E+ TA LR I
Sbjct 156 VSDGTQEGVLTAEFPLEAAEPILELDSRRTERKVVLSADTGEGSDAPEEETAALRSTI 213
> dre:100150342 si:dkey-177p2.10; si:dkey-177p2.16
Length=355
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 5/100 (5%)
Query 11 ENPEKMVKV--TKEDLEK-LEIKEEEQEELLKYISDDSDEDEEALTKKLEAFEPLLQMDD 67
E EK +K+ T+ ++K ++ KE+E + + IS SD +A+ +AF L+++ +
Sbjct 181 ERKEKELKLSDTQTKVKKTIQAKEKELQNMKMDISSHSDSALKAIKNSKKAFAELVKLIE 240
Query 68 RFPDTVI--MVGVPLVGADRLEKLTAVLRKKIADELSRKG 105
+ VI + D EK+ L+ +IAD R+G
Sbjct 241 KKSSEVIEKIKAQEKADLDEGEKIQEKLKGEIADLKKREG 280
> dre:541505 cep68, si:ch211-101n13.4, si:zc101n13.4, zgc:112945;
centrosomal protein 68
Length=650
Score = 28.1 bits (61), Expect = 7.2, Method: Composition-based stats.
Identities = 21/90 (23%), Positives = 41/90 (45%), Gaps = 14/90 (15%)
Query 6 HVFGAENPEKMVKVTKEDLEKLEIKEEEQ----------EELLKYISDDSDEDEEALTKK 55
H+ E P +++ +DL + ++E E EL + D +E +E+L +
Sbjct 504 HLLKTEEPPTSIQMISQDLNRNNLREVEAIMDQLRGLSVSELQGTMQTDENETKESLMQH 563
Query 56 LEAF----EPLLQMDDRFPDTVIMVGVPLV 81
++AF E L+Q + + V ++ P V
Sbjct 564 IQAFCSNLEVLIQWLHKVVEKVELLSPPTV 593
> eco:b3987 rpoB, ECK3978, ftsR, groN, JW3950, mbrD?, nitB, rif,
ron, sdgB, stl, stv, tabD, tabG; RNA polymerase, beta subunit
(EC:2.7.7.6); K03043 DNA-directed RNA polymerase subunit
beta [EC:2.7.7.6]
Length=1342
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 1/82 (1%)
Query 7 VFGAENPEKMVKVTKEDLEKLEIKEEEQEELLKYISDDSDEDEEALTKKLEA-FEPLLQM 65
V G EK+ K+ ++ +L + +EE++ L+ +++ DE + KKLEA + Q
Sbjct 980 VAGGVEAEKLDKLPRDRWLELGLTDEEKQNQLEQLAEQYDELKHEFEKKLEAKRRKITQG 1039
Query 66 DDRFPDTVIMVGVPLVGADRLE 87
DD P + +V V L R++
Sbjct 1040 DDLAPGVLKIVKVYLAVKRRIQ 1061
Lambda K H
0.312 0.136 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2047611720
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40