bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2341_orf1
Length=268
Score E
Sequences producing significant alignments: (Bits) Value
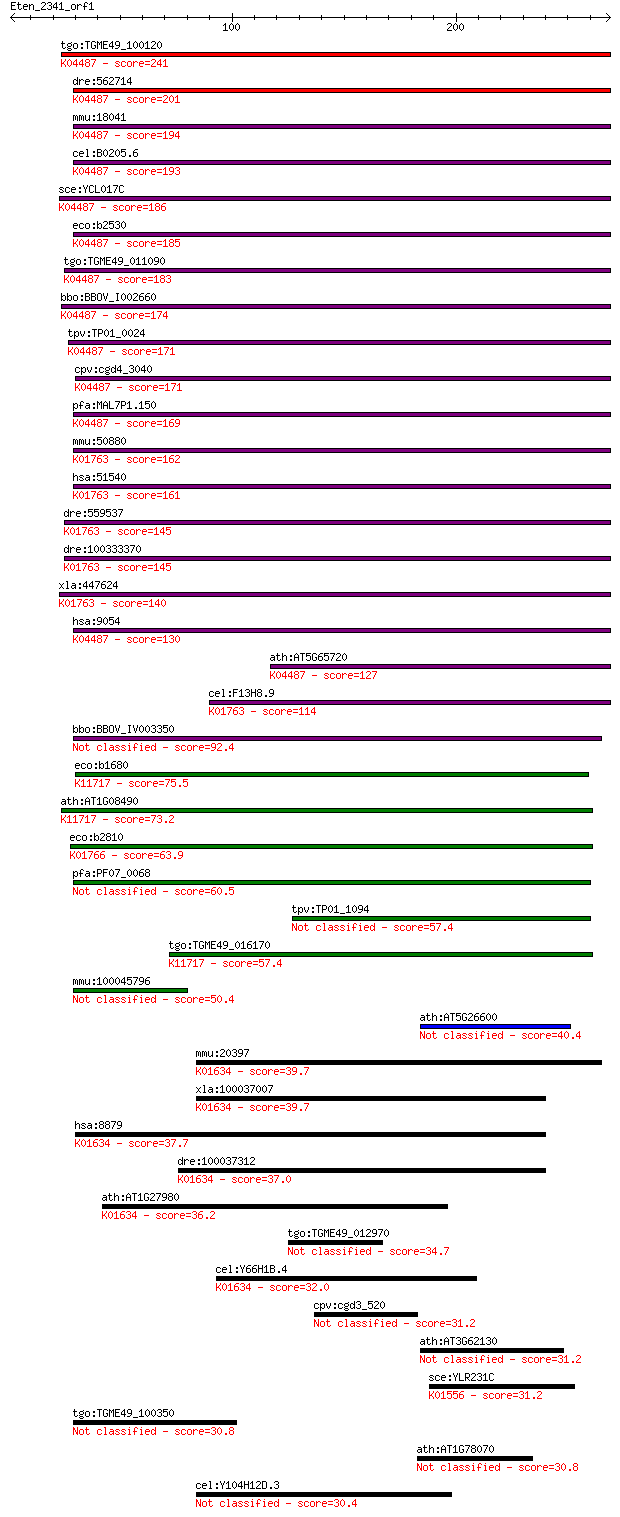
tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);... 241 2e-63
dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation 1... 201 3e-51
mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation ge... 194 4e-49
cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase ... 193 5e-49
sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-... 186 7e-47
eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine de... 185 2e-46
tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);... 183 7e-46
bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);... 174 3e-43
tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfuras... 171 3e-42
cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487... 171 3e-42
pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-); K0... 169 1e-41
mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2; ... 162 1e-39
hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);... 161 2e-39
dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16... 145 1e-34
dre:100333370 selenocysteine lyase-like; K01763 selenocysteine... 145 1e-34
xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);... 140 4e-33
hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog (S... 130 4e-30
ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/ ... 127 6e-29
cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase ... 114 3e-25
bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7) 92.4 1e-18
eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfur... 75.5 2e-13
ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE... 73.2 1e-12
eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desu... 63.9 5e-10
pfa:PF07_0068 cysteine desulfurase, putative (EC:4.4.1.-) 60.5 6e-09
tpv:TP01_1094 cysteine desulfurase 57.4 5e-08
tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);... 57.4 6e-08
mmu:100045796 cysteine desulfurase, mitochondrial-like 50.4 6e-06
ath:AT5G26600 catalytic/ pyridoxal phosphate binding 40.4 0.006
mmu:20397 Sgpl1, AI428538, D10Xrf456, S1PL, Spl; sphingosine p... 39.7 0.012
xla:100037007 sgpl1; sphingosine-1-phosphate lyase 1 (EC:4.1.2... 39.7 0.013
hsa:8879 SGPL1, FLJ13811, KIAA1252, SPL; sphingosine-1-phospha... 37.7 0.046
dre:100037312 sgpl1, zgc:162153; sphingosine-1-phosphate lyase... 37.0 0.085
ath:AT1G27980 DPL1; DPL1; carboxy-lyase/ catalytic/ pyridoxal ... 36.2 0.13
tgo:TGME49_012970 protein kinase, putative (EC:2.7.11.17 3.1.3.2) 34.7 0.39
cel:Y66H1B.4 spl-1; Sphingosine Phosphate Lyase family member ... 32.0 2.4
cpv:cgd3_520 cysteine-rich extracellular protein with a signal... 31.2 4.0
ath:AT3G62130 epimerase-related 31.2 4.1
sce:YLR231C BNA5; Bna5p (EC:3.7.1.3); K01556 kynureninase [EC:... 31.2 4.6
tgo:TGME49_100350 molybdopterin cofactor sulfurase, putative (... 30.8 5.2
ath:AT1G78070 hypothetical protein 30.8 5.6
cel:Y104H12D.3 hypothetical protein 30.4 7.7
> tgo:TGME49_100120 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=851
Score = 241 bits (615), Expect = 2e-63, Method: Compositional matrix adjust.
Identities = 124/251 (49%), Positives = 159/251 (63%), Gaps = 7/251 (2%)
Query 24 HGADG--LYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAA 81
H +G LYLD N++T DP VF ++PF + FGNP S H G AL RE A
Sbjct 194 HNREGFPLYLDNNSSTKTDPRVFQEMAPFFESLFGNPGSAHERGRINKAALEEGRERVAG 253
Query 82 ALGAPPCSIIFTSGAT----ESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEY 137
LG PP +I FTSGAT ES+NWA+K GA + KG IVT +EHPAVL +C++
Sbjct 254 CLGVPPSTIFFTSGATGINSESLNWAIKCGATAQSRKGLDR-HIVTTRIEHPAVLEICKF 312
Query 138 LRDEEGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARR 197
L ++ GF++ CPV G ++L A LL A+T +S+P AN E+G VQPI +V + R
Sbjct 313 LEEDHGFQVTFCPVDCFGFVNLEALSRLLRAETAFVSVPHANAEIGAVQPIEKVAMIVRE 372
Query 198 ICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGINLPPLILG 257
P AL+H D SQ++GK+ V LG DL+T+AGHK+YAPKGVGALY+ L PL+ G
Sbjct 373 HAPHALLHVDCSQSLGKIPVNIPQLGADLVTIAGHKIYAPKGVGALYVGSRACLGPLLHG 432
Query 258 GGQESGLRGGT 268
GGQE LRGGT
Sbjct 433 GGQERRLRGGT 443
> dre:562714 nfs1, fb50g03, wu:fb50g03; NFS1 nitrogen fixation
1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=451
Score = 201 bits (510), Expect = 3e-51, Method: Compositional matrix adjust.
Identities = 106/243 (43%), Positives = 149/243 (61%), Gaps = 9/243 (3%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPP 87
LY+D+ ATTP+DP V A+ P+ +GNP S+ HA G ++ A+ AR+ A +GA P
Sbjct 53 LYMDFQATTPMDPRVLDAMLPYQVNYYGNPHSRTHAYGWESESAMEKARKQVAGLIGADP 112
Query 88 CSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELI 147
I+FTSGATES N ++K A+ + K I+T +EH VL C L + EGF++
Sbjct 113 REIVFTSGATESNNMSIKGVARFYKAK---KMHIITTQIEHKCVLDSCRVL-ETEGFDIT 168
Query 148 LCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHAD 207
PV S+GLIDL + + DT L+SI N E+GV QP++E+ + R + H D
Sbjct 169 YLPVKSNGLIDLKQLEDTIRPDTSLVSIMAINNEIGVKQPVKEIGHLCR--SKNVFFHTD 226
Query 208 ASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE--GINLPPLILGGGQESGLR 265
A+QA+GK+ V VDL++++ HK+Y PKGVGAL++R + L PL GGGQE GLR
Sbjct 227 AAQAVGKIPVDVTDWKVDLMSISAHKIYGPKGVGALFVRRRPRVRLEPLQSGGGQERGLR 286
Query 266 GGT 268
GT
Sbjct 287 SGT 289
> mmu:18041 Nfs1, AA987187, m-Nfs1, m-Nfsl; nitrogen fixation
gene 1 (S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=459
Score = 194 bits (492), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 108/245 (44%), Positives = 149/245 (60%), Gaps = 13/245 (5%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPP 87
LY+D ATTP+DP V A+ P+L +GNP S+ HA G ++ A+ AR+ A+ +GA P
Sbjct 61 LYMDVQATTPLDPRVLDAMLPYLVNYYGNPHSRTHAYGWESEAAMERARQQVASLIGADP 120
Query 88 CSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELI 147
IIFTSGATES N A+K A+ R + +VT EH VL C L + EGF +
Sbjct 121 REIIFTSGATESNNIAIKGVARFYRSRKK---HLVTTQTEHKCVLDSCRSL-EAEGFRVT 176
Query 148 LCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALV--H 205
PV SG+IDL +A + DT L+S+ N E+GV QPI E+ +IC V H
Sbjct 177 YLPVQKSGIIDLKELEAAIQPDTSLVSVMTVNNEIGVKQPIAEI----GQICSSRKVYFH 232
Query 206 ADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE--GINLPPLILGGGQESG 263
DA+QA+GK+ + + +DL++++GHK+Y PKGVGA+YIR + + L GGGQE G
Sbjct 233 TDAAQAVGKIPLDVNDMKIDLMSISGHKLYGPKGVGAIYIRRRPRVRVEALQSGGGQERG 292
Query 264 LRGGT 268
+R GT
Sbjct 293 MRSGT 297
> cel:B0205.6 hypothetical protein; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=412
Score = 193 bits (491), Expect = 5e-49, Method: Compositional matrix adjust.
Identities = 102/243 (41%), Positives = 144/243 (59%), Gaps = 8/243 (3%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPP 87
+YLD AT P+DP V A+ P++ FGNP S+ H+ G A + ARE A + A P
Sbjct 13 IYLDVQATAPMDPRVVDAMLPYMINDFGNPHSRTHSYGWKAEEGVEQAREHVANLIKADP 72
Query 88 CSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELI 147
IIFTSGATES N A+K AK ++ G + I+T EH VL C YL +E GF++
Sbjct 73 RDIIFTSGATESNNLAIKGVAKFRKQSGKN--HIITLQTEHKCVLDSCRYLENE-GFKVT 129
Query 148 LCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHAD 207
PV G++D+ +TA+T L+SI N E+GV+QPI+++ + R H D
Sbjct 130 YLPVDKGGMVDMEQLTQSITAETCLVSIMFVNNEIGVMQPIKQIGELCR--SKGVYFHTD 187
Query 208 ASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE--GINLPPLILGGGQESGLR 265
A+QA GKV + + +DL++++ HK+Y PKG GALY+R + + + GGGQE GLR
Sbjct 188 AAQATGKVPIDVNEMKIDLMSISAHKIYGPKGAGALYVRRRPRVRIEAQMSGGGQERGLR 247
Query 266 GGT 268
GT
Sbjct 248 SGT 250
> sce:YCL017C NFS1, SPL1; Cysteine desulfurase involved in iron-sulfur
cluster (Fe/S) biogenesis; required for the post-transcriptional
thio-modification of mitochondrial and cytoplasmic
tRNAs; essential protein located predominantly in mitochondria
(EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=497
Score = 186 bits (472), Expect = 7e-47, Method: Compositional matrix adjust.
Identities = 101/249 (40%), Positives = 140/249 (56%), Gaps = 9/249 (3%)
Query 23 GHGADGLYLDYNATTPVDPAVFAALSPFLQERFGNP-SSKHAVGVDAAGALRAAREAAAA 81
G G +YLD ATTP DP V + F +GNP S+ H+ G + A+ AR A
Sbjct 94 GFGTRPIYLDMQATTPTDPRVLDTMLKFYTGLYGNPHSNTHSYGWETNTAVENARAHVAK 153
Query 82 ALGAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDE 141
+ A P IIFTSGATES N LK + R + I+T EH VL + +
Sbjct 154 MINADPKEIIFTSGATESNNMVLKG---VPRFYKKTKKHIITTRTEHKCVLEAARAMM-K 209
Query 142 EGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPD 201
EGFE+ V GLIDL + + DT L+S+ N E+GV+QPI+E+ ++ R+
Sbjct 210 EGFEVTFLNVDDQGLIDLKELEDAIRPDTCLVSVMAVNNEIGVIQPIKEIGAICRK--NK 267
Query 202 ALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE--GINLPPLILGGG 259
H DA+QA GK+ + + +DLL+++ HK+Y PKG+GA+Y+R + L PL+ GGG
Sbjct 268 IYFHTDAAQAYGKIHIDVNEMNIDLLSISSHKIYGPKGIGAIYVRRRPRVRLEPLLSGGG 327
Query 260 QESGLRGGT 268
QE GLR GT
Sbjct 328 QERGLRSGT 336
> eco:b2530 iscS, ECK2527, JW2514, nuvC, yfhO, yzzO; cysteine
desulfurase (tRNA sulfurtransferase), PLP-dependent (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=404
Score = 185 bits (469), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 105/245 (42%), Positives = 143/245 (58%), Gaps = 11/245 (4%)
Query 29 LYLDYNATTPVDPAVFAALSPFL--QERFGNPSSK-HAVGVDAAGALRAAREAAAAALGA 85
+YLDY+ATTPVDP V + F+ FGNP+S+ H G A A+ AR A +GA
Sbjct 5 IYLDYSATTPVDPRVAEKMMQFMTMDGTFGNPASRSHRFGWQAEEAVDIARNQIADLVGA 64
Query 86 PPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFE 145
P I+FTSGATES N A+K A + KG I+T EH AVL C L + EGFE
Sbjct 65 DPREIVFTSGATESDNLAIKGAANFYQKKGK---HIITSKTEHKAVLDTCRQL-EREGFE 120
Query 146 LILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVH 205
+ +G+IDL +A + DTIL+SI N E+GVVQ I + + R + H
Sbjct 121 VTYLAPQRNGIIDLKELEAAMRDDTILVSIMHVNNEIGVVQDIAAIGEMCR--ARGIIYH 178
Query 206 ADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREG--INLPPLILGGGQESG 263
DA+Q++GK+ + L VDL++ +GHK+Y PKG+GALY+R + + + GGG E G
Sbjct 179 VDATQSVGKLPIDLSQLKVDLMSFSGHKIYGPKGIGALYVRRKPRVRIEAQMHGGGHERG 238
Query 264 LRGGT 268
+R GT
Sbjct 239 MRSGT 243
> tgo:TGME49_011090 cysteine desulfurase, putative (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=487
Score = 183 bits (464), Expect = 7e-46, Method: Compositional matrix adjust.
Identities = 107/258 (41%), Positives = 149/258 (57%), Gaps = 19/258 (7%)
Query 25 GADGLYLDYNATTPVDPAVFAALSPFLQERFGNP-SSKHAVGVDAAGALRAAREAAAAAL 83
G+ +YLD ATT DP V A+ PFL ++FGNP SS HAVG +A A+ AR+ A L
Sbjct 62 GSRPVYLDNQATTVQDPRVTDAMLPFLFDKFGNPHSSSHAVGWEADAAVEKARKQVAHLL 121
Query 84 GAPPC---SIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRD 140
G IIFTSGATES N ALK + + ++T +EH L+ C L+
Sbjct 122 GLDASRAREIIFTSGATESNNLALKGATRAA---ARARRHVITTQLEHKCALQCCRMLQL 178
Query 141 E-------EGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVS 193
E G ++ PV + GL+DL + + DT+L+S+ N E+GVVQ + E+
Sbjct 179 EFSESQGARGCDVTYLPVKTDGLVDLEELEKAIRPDTLLVSVMFVNNEIGVVQNLEEIGK 238
Query 194 VARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREG---IN 250
+ +R D L H DA+Q GK+ + +G+DLL+++ HK+Y PKG+GAL++R +
Sbjct 239 ICKR--HDILFHTDAAQGAGKLPIDVDEMGIDLLSLSSHKIYGPKGIGALFVRAKNPRVR 296
Query 251 LPPLILGGGQESGLRGGT 268
L PLI GGGQE GLR GT
Sbjct 297 LQPLIDGGGQERGLRSGT 314
> bbo:BBOV_I002660 19.m02112; cysteine desulfurase (EC:2.8.1.7);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=490
Score = 174 bits (441), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 99/253 (39%), Positives = 139/253 (54%), Gaps = 11/253 (4%)
Query 24 HGADGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAA 82
H D +Y+DY ATTP+DP V + P+L FGNP S+ H+ G DA A+ AR A+
Sbjct 80 HHNDRVYMDYQATTPLDPRVLDKMMPYLTHSFGNPHSRTHSFGWDAEHAVEEARSQVASL 139
Query 83 LGAPPCSIIFTSGATESINWALKSGAKL-----KRLKGPSPGRIVTCAMEHPAVLRVCEY 137
L SIIFTSGATES N ALK + + ++T ++H VL+ C
Sbjct 140 LNCDSRSIIFTSGATESNNLALKGITEYYSCLQHNDQSKIKDHLITSQIDHKCVLQTCRQ 199
Query 138 LRDEEGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARR 197
L + G+ + SG+I A +T T + SI N E+G +Q I + + R
Sbjct 200 L-ENRGYTVTYLKPDKSGIIKPEDVAAAITPRTFMCSIIHLNNEIGTIQNIAAIGKICRE 258
Query 198 ICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYI--REGINLPPLI 255
+ H D +Q+ GK+ + +L VDL++++GHK+Y PKGVGALY+ + I L P+I
Sbjct 259 --KGVVFHTDGAQSFGKIPIDLSALNVDLMSISGHKIYGPKGVGALYVGRKPRIRLRPII 316
Query 256 LGGGQESGLRGGT 268
GGGQE GLR GT
Sbjct 317 DGGGQERGLRSGT 329
> tpv:TP01_0024 cysteine desulfurase; K04487 cysteine desulfurase
[EC:2.8.1.7]
Length=448
Score = 171 bits (433), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 99/250 (39%), Positives = 141/250 (56%), Gaps = 11/250 (4%)
Query 27 DGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGA 85
+ +YLD ATT VDP V A+ P+L FGNP S+ H+ G +A A+ AR A +
Sbjct 41 NRVYLDNQATTCVDPRVLDAMMPYLTHAFGNPHSRTHSYGWEAEKAVETARADIANLINC 100
Query 86 PPCSIIFTSGATESINWALKSGAKLKRLKGPSPGR-----IVTCAMEHPAVLRVCEYLRD 140
++IFTSGATES N A+K SPG+ ++T +EH VL+ C L +
Sbjct 101 ESKNVIFTSGATESNNLAIKGSKSFYGRLVESPGKSKKNHVITTQIEHKCVLQCCRQLEN 160
Query 141 EEGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICP 200
E G+ + G+I + + +T L S+ N E+GV+Q I E+ V +
Sbjct 161 E-GYSVTYLKPDKYGMILPEEVRKNIRPETFLCSVIHVNNEIGVIQDIAEIGKVCKE--H 217
Query 201 DALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREG--INLPPLILGG 258
+ H DA+Q+ GK+ + +L VDLL+++GHK+Y PKGVGAL++R I L P+I GG
Sbjct 218 KVIFHTDAAQSFGKLPIDLKNLEVDLLSISGHKIYGPKGVGALFVRTKPRIRLQPIIDGG 277
Query 259 GQESGLRGGT 268
GQE GLR GT
Sbjct 278 GQERGLRSGT 287
> cpv:cgd4_3040 NifS-like protein; cysteine desulfurase ; K04487
cysteine desulfurase [EC:2.8.1.7]
Length=438
Score = 171 bits (432), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 95/243 (39%), Positives = 133/243 (54%), Gaps = 7/243 (2%)
Query 30 YLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPPC 88
Y DY ATTPVDP V + PF E+FGN S+ H G +A A+ AR A + P
Sbjct 37 YFDYQATTPVDPRVLDKMMPFFTEKFGNSHSRTHGYGWEAEEAVENARTNIANLIKCLPK 96
Query 89 SIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELIL 148
IIFTSGATES N ++ + I+T +EH VL L + +GF +
Sbjct 97 EIIFTSGATESNNTIIRGVCDIYGDIENKKNHIITTQIEHKCVLSTLREL-ELKGFRVTY 155
Query 149 CPVLSSGLIDLGAF-KALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHAD 207
V + GLI L K+++ +TIL SI N E+GV+QP+ + + ++ + L H+D
Sbjct 156 LKVNNKGLISLEELEKSIIPGETILASIMHVNNEIGVIQPMNLIGEICKKY--NVLFHSD 213
Query 208 ASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREG--INLPPLILGGGQESGLR 265
+Q +GK+ + D L+++ HKVY PKG+GA YIR + PLI GGGQE G+R
Sbjct 214 VAQGLGKINIDVDKWNADFLSLSAHKVYGPKGIGAFYIRSKPRRRIKPLIFGGGQERGMR 273
Query 266 GGT 268
GT
Sbjct 274 SGT 276
> pfa:MAL7P1.150 cysteine desulfurase, putative (EC:4.4.1.-);
K04487 cysteine desulfurase [EC:2.8.1.7]
Length=553
Score = 169 bits (427), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 93/245 (37%), Positives = 134/245 (54%), Gaps = 8/245 (3%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAAL-GAP 86
YLD ATT +DP V + P++ +GN S+ H G ++ A+ AR + G
Sbjct 147 FYLDSQATTMIDPRVLDKMLPYMTYIYGNAHSRNHFFGWESEKAVEDARTNLLNLINGKN 206
Query 87 PCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFEL 146
IIFTSGATES N AL I+T +EH +L+ C +L+ +GFE+
Sbjct 207 NKEIIFTSGATESNNLALIGICTYYNKLNKQKNHIITSQIEHKCILQTCRFLQ-TKGFEV 265
Query 147 ILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHA 206
++GL+ L K + +TI+ S N E+GV+Q I + ++ + + L H
Sbjct 266 TYLKPDTNGLVKLDDIKNSIKDNTIMASFIFVNNEIGVIQDIENIGNLCKE--KNILFHT 323
Query 207 DASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE---GINLPPLILGGGQESG 263
DASQA GKV + + +DL++++GHK+Y PKG+GALYI+ I L LI GGGQE G
Sbjct 324 DASQAAGKVPIDVQKMNIDLMSMSGHKLYGPKGIGALYIKRKKPNIRLNALIHGGGQERG 383
Query 264 LRGGT 268
LR GT
Sbjct 384 LRSGT 388
> mmu:50880 Scly, 9830169H08, A930015N15Rik, SCL, Scly1, Scly2;
selenocysteine lyase (EC:4.4.1.16); K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=432
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 104/270 (38%), Positives = 147/270 (54%), Gaps = 35/270 (12%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALGAPPC 88
+Y+DYNATTP++P V A++ ++E +GNPSS + G A + AAR + A +G P
Sbjct 20 VYMDYNATTPLEPEVIQAVTEAMKEAWGNPSSSYVSGRKAKDIINAARASLAKMIGGKPQ 79
Query 89 SIIFTSGATESINWALKSGAK----LKRLKG-------PSPG---RIVTCAMEHPAVLRV 134
IIFTSG TES N + S + + LKG P G +TC +EH ++
Sbjct 80 DIIFTSGGTESNNLVIHSMVRCFHEQQTLKGNMVDQHSPEEGTRPHFITCTVEHDSIRLP 139
Query 135 CEYLRDEEGFELILCPVLS-SGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVS 193
E+L + + E+ PV +G ++ A + T L++I LAN E GV+ P+ E
Sbjct 140 LEHLVENQMAEVTFVPVSKVNGQAEVEDILAAVRPTTCLVTIMLANNETGVIMPVSE--- 196
Query 194 VARRI-----------CPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGA 242
++RRI P LVH DA+QA+GK V LGVD LT+ GHK Y P+ +GA
Sbjct 197 ISRRIKALNQIRAASGLPRVLVHTDAAQALGKRRVDVEDLGVDFLTIVGHKFYGPR-IGA 255
Query 243 LYIREGIN----LPPLILGGGQESGLRGGT 268
LY+R G+ L P++ GGGQE R GT
Sbjct 256 LYVR-GVGKLTPLYPMLFGGGQEWNFRPGT 284
> hsa:51540 SCLY, SCL, hSCL; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=453
Score = 161 bits (408), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 102/267 (38%), Positives = 143/267 (53%), Gaps = 29/267 (10%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALGAPPC 88
+Y+DYNATTP++P V A++ + E +GNPSS ++ G A + AARE+ A +G P
Sbjct 40 VYMDYNATTPLEPEVIQAMTKAMWEAWGNPSSPYSAGRKAKDIINAARESLAKMIGGKPQ 99
Query 89 SIIFTSGATESINWALKS---------------GAKLKRLKGPSPGRIVTCAMEHPAVLR 133
IIFTSG TES N + S G +KG P +T ++EH ++
Sbjct 100 DIIFTSGGTESNNLVIHSVVKHFHANQTSKGHTGGHHSPVKGAKP-HFITSSVEHDSIRL 158
Query 134 VCEYLRDEEGFELILCPVLS-SGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVV 192
E+L +E+ + PV SG ++ A + T L++I LAN E G+V P+ E+
Sbjct 159 PLEHLVEEQVAAVTFVPVSKVSGQAEVDDILAAVRPTTRLVTIMLANNETGIVMPVPEIS 218
Query 193 SVARRI--------CPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALY 244
+ + P LVH DA+QA+GK V LGVD LT+ GHK Y P+ +GALY
Sbjct 219 QRIKALNQERVAAGLPPILVHTDAAQALGKQRVDVEDLGVDFLTIVGHKFYGPR-IGALY 277
Query 245 IR---EGINLPPLILGGGQESGLRGGT 268
IR E L P++ GGGQE R GT
Sbjct 278 IRGLGEFTPLYPMLFGGGQERNFRPGT 304
> dre:559537 scly, zgc:171514; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=450
Score = 145 bits (367), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 91/271 (33%), Positives = 137/271 (50%), Gaps = 29/271 (10%)
Query 25 GADGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALG 84
G D +Y+DYNATTP DP V ++ L + +GNPSS + G+ A + +R+A A +G
Sbjct 33 GPDRIYMDYNATTPADPEVIRVVTDALMDAWGNPSSNYLPGLKARDIIYHSRDAIARMVG 92
Query 85 APPCSIIFTSGATESINWALK----------------SGAKLKRLKGPSPGRIVTCAMEH 128
IIFTSG TE+ N + +K +G P ++ +EH
Sbjct 93 GKAADIIFTSGGTEANNLVFHTAVEHFKKSMISAEGSTSSKQTNGRGSLP-HVIISNVEH 151
Query 129 PAVLRVCEYLRDEEGFELILCPVLS-SGLIDLGAFKALLTADTILISIPLANGEVGVVQP 187
++ E L E ++ PV + +++ A + T L+SI LAN E G++ P
Sbjct 152 DSIKLTAENLLKEGKADVTFVPVSKVTARVEVEDVIAAIRPTTCLVSIMLANNETGIIMP 211
Query 188 IREVVSVARRI-------CPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGV 240
I+++ + P L+H DA+QA+GK+ V LGVD LT+ GHK Y P+
Sbjct 212 IKDICQRVNEVNKQRAASAPRILLHTDAAQAIGKIRVDAHELGVDYLTIVGHKFYGPR-T 270
Query 241 GALYIRE-GINLP--PLILGGGQESGLRGGT 268
GAL++ + G + P P+ GGGQE R GT
Sbjct 271 GALFVNDPGKSTPVYPMFFGGGQERNFRPGT 301
> dre:100333370 selenocysteine lyase-like; K01763 selenocysteine
lyase [EC:4.4.1.16]
Length=450
Score = 145 bits (367), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 91/271 (33%), Positives = 137/271 (50%), Gaps = 29/271 (10%)
Query 25 GADGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALG 84
G D +Y+DYNATTP DP V ++ L + +GNPSS + G+ A + +R+A A +G
Sbjct 33 GPDRIYMDYNATTPADPEVIRVVTDALMDAWGNPSSNYLPGLKARDIIYHSRDAIARMVG 92
Query 85 APPCSIIFTSGATESINWALK----------------SGAKLKRLKGPSPGRIVTCAMEH 128
IIFTSG TE+ N + +K +G P ++ +EH
Sbjct 93 GKAADIIFTSGGTEANNLVFHTAVEHFKKSMISAEGSTSSKQTNGRGSLP-HVIISNVEH 151
Query 129 PAVLRVCEYLRDEEGFELILCPVLS-SGLIDLGAFKALLTADTILISIPLANGEVGVVQP 187
++ E L E ++ PV + +++ A + T L+SI LAN E G++ P
Sbjct 152 DSIKLTAENLLKEGKADVTFVPVSKVTARVEVEDVIAAIRPTTCLVSIMLANNETGIIMP 211
Query 188 IREVVSVARRI-------CPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGV 240
I+++ + P L+H DA+QA+GK+ V LGVD LT+ GHK Y P+
Sbjct 212 IKDICQRVNEVNKQRAASAPRILLHTDAAQAIGKIRVDAHELGVDYLTIVGHKFYGPR-T 270
Query 241 GALYIRE-GINLP--PLILGGGQESGLRGGT 268
GAL++ + G + P P+ GGGQE R GT
Sbjct 271 GALFVNDPGKSTPVYPMFFGGGQERNFRPGT 301
> xla:447624 scly, MGC85597; selenocysteine lyase (EC:4.4.1.16);
K01763 selenocysteine lyase [EC:4.4.1.16]
Length=426
Score = 140 bits (354), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 98/270 (36%), Positives = 141/270 (52%), Gaps = 28/270 (10%)
Query 23 GHGADGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAA 82
H +YLDYNATTP V ++ LQE +GNPSS + G A + AR A
Sbjct 11 NHLNHKIYLDYNATTPPAREVVGTVAEALQEAWGNPSSSYTAGCKAKELIDTARARIAKM 70
Query 83 LGAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGR-----------IVTCAMEHPAV 131
+G P IIFTSG TE+ N L S ++ S R I+T +EH +V
Sbjct 71 VGGKPEDIIFTSGGTEANNMVLFSA--VENFNRTSKERQNNNVDWALPHIITSNVEHDSV 128
Query 132 LRVCEYLRDEEGFELILCPVLS-SGLIDLGAFKALLTADTILISIPLANGEVGVVQPIRE 190
L+ E+ PV + +G +++ + + +T L+SI LAN E GV+ P+ E
Sbjct 129 ALPLLQLQKTHKAEITFVPVSTVTGRVEVEDVISAVRPNTCLVSIMLANNETGVIMPVGE 188
Query 191 ----VVSVARRIC----PDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGA 242
+ S++R+ P+ L+H DA+QA+GKV V LGV+ LT+ GHK Y P+ +GA
Sbjct 189 LSQCLASLSRKRSAQGLPEILLHTDAAQALGKVEVDVQELGVNYLTIVGHKFYGPR-IGA 247
Query 243 LYIREGIN----LPPLILGGGQESGLRGGT 268
LY+ G+ L P++ GGG+E R GT
Sbjct 248 LYV-GGLGHQSPLLPMLYGGGREGNFRPGT 276
> hsa:9054 NFS1, IscS, NIFS; NFS1 nitrogen fixation 1 homolog
(S. cerevisiae) (EC:2.8.1.7); K04487 cysteine desulfurase [EC:2.8.1.7]
Length=406
Score = 130 bits (328), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 85/245 (34%), Positives = 119/245 (48%), Gaps = 64/245 (26%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPP 87
LY+D ATTP+DP V A+ P+L +GNP S+ HA G ++ A+ AR+ A+ +GA P
Sbjct 59 LYMDVQATTPLDPRVLDAMLPYLINYYGNPHSRTHAYGWESEAAMERARQQVASLIGADP 118
Query 88 CSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELI 147
IIFT SGA
Sbjct 119 REIIFT------------SGAT-------------------------------------- 128
Query 148 LCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALV--H 205
S I + +A + DT L+S+ N E+GV QPI E+ RIC V H
Sbjct 129 -----ESNNIAIKELEAAIQPDTSLVSVMTVNNEIGVKQPIAEI----GRICSSRKVYFH 179
Query 206 ADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIRE--GINLPPLILGGGQESG 263
DA+QA+GK+ + + +DL++++GHK+Y PKGVGA+YIR + + L GGGQE G
Sbjct 180 TDAAQAVGKIPLDVNDMKIDLMSISGHKIYGPKGVGAIYIRRRPRVRVEALQSGGGQERG 239
Query 264 LRGGT 268
+R GT
Sbjct 240 MRSGT 244
> ath:AT5G65720 NFS1; NFS1; ATP binding / cysteine desulfurase/
transaminase; K04487 cysteine desulfurase [EC:2.8.1.7]
Length=325
Score = 127 bits (318), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 69/156 (44%), Positives = 95/156 (60%), Gaps = 9/156 (5%)
Query 117 SPGRIVTCAMEHPAVLRVCEYLRDEEGFELILCPVLSSGLIDLGAFKALLTADTILISIP 176
+ ++T EH VL C +L+ +EGFE+ PV + GL+DL + + DT L+SI
Sbjct 13 TKKHVITTQTEHKCVLDSCRHLQ-QEGFEVTYLPVKTDGLVDLEMLREAIRPDTGLVSIM 71
Query 177 LANGEVGVVQPIREVVSVARRICPDALV--HADASQAMGKVAVQPVSLGVDLLTVAGHKV 234
N E+GVVQP+ E+ IC + V H DA+QA+GK+ V V L++++ HK+
Sbjct 72 AVNNEIGVVQPMEEI----GMICKEHNVPFHTDAAQAIGKIPVDVKKWNVALMSMSAHKI 127
Query 235 YAPKGVGALYIRE--GINLPPLILGGGQESGLRGGT 268
Y PKGVGALY+R I L PL+ GGGQE GLR GT
Sbjct 128 YGPKGVGALYVRRRPRIRLEPLMNGGGQERGLRSGT 163
> cel:F13H8.9 hypothetical protein; K01763 selenocysteine lyase
[EC:4.4.1.16]
Length=328
Score = 114 bits (285), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 66/184 (35%), Positives = 101/184 (54%), Gaps = 8/184 (4%)
Query 90 IIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELILC 149
++FTSG TES NW ++ + + P I+T +EHP++L + R EE E+ +
Sbjct 9 VVFTSGGTESNNWVIEGTIRNAKKVSKLP-HIITTNIEHPSILEPLK--RREEDGEISVT 65
Query 150 PVLS---SGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDA--LV 204
V +G + + LT+DT L++I LAN + GV+QP+ E+ R +
Sbjct 66 YVSINPLTGFVTSQSILDALTSDTCLVTIMLANNDTGVLQPVSEIFQAIREKLKTNVPFL 125
Query 205 HADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGINLPPLILGGGQESGL 264
H+D +QA GK+ V SL D +TV GHK Y P+ ++ + +PP++LGG QESG
Sbjct 126 HSDVAQAAGKIPVNVRSLSADAVTVVGHKFYGPRSGALIFNPKSKRIPPMLLGGNQESGW 185
Query 265 RGGT 268
R GT
Sbjct 186 RSGT 189
> bbo:BBOV_IV003350 21.m02766; cysteine desulfurase (EC:2.8.1.7)
Length=423
Score = 92.4 bits (228), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 73/242 (30%), Positives = 113/242 (46%), Gaps = 12/242 (4%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGN-PSSKHAVGVDAAGALRAAREAAAAALGAPP 87
+YLD ATT V A++ + N S++ + + AR A A + AP
Sbjct 73 IYLDNAATTQKPYCVIEAITNYYASPCSNVHRSQYQLATAVSNQYEDARTAIAKFINAPS 132
Query 88 C-SIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAM-EHPAVLRVCEYLRDEEGFE 145
+IIFTSGAT+SIN + PG V + EH + + + L +
Sbjct 133 ARNIIFTSGATDSINLVANAWG----YTHIRPGDTVLVPLSEHNSNILPWKLLEQKNNSN 188
Query 146 LILCPVLSSGLIDLGAFKALLTADTI-LISIPLANGEVGVVQPIREVVSVARRICPDALV 204
+ + + G +DL +K L+ + LISI A+ +GVVQ ++ +++ A ALV
Sbjct 189 VHFVKLNTDGTLDLDDYKRQLSTGKVRLISIAHASNVLGVVQDLKSIIATAHE--HGALV 246
Query 205 HADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGI--NLPPLILGGGQES 262
DA Q + V + L D L +GHKVY P G+G LY ++ + ++ P GGG
Sbjct 247 LVDACQTLAHVNIDVQQLNCDFLVASGHKVYGPTGIGFLYAKQEVIEHMSPYKGGGGMVK 306
Query 263 GL 264
L
Sbjct 307 NL 308
> eco:b1680 sufS, csdB, ECK1676, JW1670, ynhB; cysteine desulfurase,
stimulated by SufE; selenocysteine lyase, PLP-dependent
(EC:4.4.1.16); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=406
Score = 75.5 bits (184), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 62/233 (26%), Positives = 100/233 (42%), Gaps = 9/233 (3%)
Query 30 YLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAAAAALGAPPC 88
YLD A+ V A + F + + H + A + R+ A+ + A
Sbjct 26 YLDSAASAQKPSQVIDAEAEFYRHGYAAVHRGIHTLSAQATEKMENVRKRASLFINARSA 85
Query 89 S-IIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELI 147
++F G TE IN S G + I+ MEH A + + L G EL
Sbjct 86 EELVFVRGTTEGINLVANSWGNSNVRAGDN---IIISQMEHHANIVPWQMLCARVGAELR 142
Query 148 LCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHAD 207
+ P+ G + L L T L++I + +G P+ E++++A + LV D
Sbjct 143 VIPLNPDGTLQLETLPTLFDEKTRLLAITHVSNVLGTENPLAEMITLAHQHGAKVLV--D 200
Query 208 ASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGI--NLPPLILGG 258
+QA+ V +L D +GHK+Y P G+G LY++E + +PP GG
Sbjct 201 GAQAVMHHPVDVQALDCDFYVFSGHKLYGPTGIGILYVKEALLQEMPPWEGGG 253
> ath:AT1G08490 CPNIFS; CPNIFS (CHLOROPLASTIC NIFS-LIKE CYSTEINE
DESULFURASE); cysteine desulfurase/ selenocysteine lyase/
transaminase (EC:2.8.1.7); K11717 cysteine desulfurase / selenocysteine
lyase [EC:2.8.1.7 4.4.1.16]
Length=463
Score = 73.2 bits (178), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 69/243 (28%), Positives = 111/243 (45%), Gaps = 14/243 (5%)
Query 24 HGADGLYLDYNATTPVDPAVFAALSPFLQERFGNPSSK---HAVGVDAAGALRAAREAAA 80
+G+ +YLD AT+ AV AL + + F N + H + A AR+ A
Sbjct 69 NGSKLVYLDSAATSQKPAAVLDALQNYYE--FYNSNVHRGIHYLSAKATDEFELARKKVA 126
Query 81 AALGAPPC-SIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLR 139
+ A I+FT ATE+IN S L LK P I+T A EH + + + +
Sbjct 127 RFINASDSREIVFTRNATEAINLVAYSWG-LSNLK-PGDEVILTVA-EHHSCIVPWQIVS 183
Query 140 DEEGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRIC 199
+ G L + + D+ + L++ T L+++ + + PI E+V A +
Sbjct 184 QKTGAVLKFVTLNEDEVPDINKLRELISPKTKLVAVHHVSNVLASSLPIEEIVVWAHDVG 243
Query 200 PDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGI--NLPPLILG 257
LV DA Q++ + V L D L + HK+ P G+G LY + + ++PP LG
Sbjct 244 AKVLV--DACQSVPHMVVDVQKLNADFLVASSHKMCGPTGIGFLYGKSDLLHSMPPF-LG 300
Query 258 GGQ 260
GG+
Sbjct 301 GGE 303
> eco:b2810 csdA, ECK2806, JW2781, ygdJ; cysteine sulfinate desulfinase
(EC:4.4.1.-); K01766 cysteine sulfinate desulfinase
[EC:4.4.1.-]
Length=401
Score = 63.9 bits (154), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 67/237 (28%), Positives = 103/237 (43%), Gaps = 10/237 (4%)
Query 28 GLYLDYNATTPVDPAVFAALSPFLQERFGN-PSSKHAVGVDAAGALRAAREAAAAALGAP 86
G+YLD AT AV A F GN S+ A AARE A L AP
Sbjct 20 GVYLDSAATALKPEAVVEATQQFYSLSAGNVHRSQFAEAQRLTARYEAAREKVAQLLNAP 79
Query 87 P-CSIIFTSGATESINWALKSGAKLKRLKGPSPG-RIVTCAMEHPAVLRVCEYLRDEEGF 144
+I++T G TESIN + A+ RL+ PG I+ EH A L + + G
Sbjct 80 DDKTIVWTRGTTESINMVAQCYAR-PRLQ---PGDEIIVSVAEHHANLVPWLMVAQQTGA 135
Query 145 ELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALV 204
+++ P+ + L D+ L+T + ++++ + G + ++ A +V
Sbjct 136 KVVKLPLNAQRLPDVDLLPELITPRSRILALGQMSNVTGGCPDLARAITFAH--SAGMVV 193
Query 205 HADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIR-EGINLPPLILGGGQ 260
D +Q L +D +GHK+Y P G+G LY + E + LGGG+
Sbjct 194 MVDGAQGAVHFPADVQQLDIDFYAFSGHKLYGPTGIGVLYGKSELLEAMSPWLGGGK 250
> pfa:PF07_0068 cysteine desulfurase, putative (EC:4.4.1.-)
Length=546
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 56/243 (23%), Positives = 104/243 (42%), Gaps = 14/243 (5%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSS-KHAVGVDAAGALRAAREAAAAALGAPP 87
+Y D ATT V +S F ++ N + + +A RE +
Sbjct 81 IYFDSAATTHKPSCVIEKMSEFYKKENSNIHRGIYKLSHNATNNYEKVRETIKEYINCEK 140
Query 88 C-SIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPA-VLRVCEYLRDEEGFE 145
+IIFT+G+T +N K + K + MEH + ++ EY+ E+
Sbjct 141 NDNIIFTNGSTYGLNVVCKMMIEEIIKKEEDEIYL--SYMEHHSNIIPWQEYINKEKKGR 198
Query 146 LILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVH 205
+ P+ SG I++ + + +T +ISI A+ +G +Q I +++ + + P ++
Sbjct 199 IKYVPLNKSGYINIKKLISNMNINTKVISICHASNVIGNIQNIEKIIKKIKNVYPHIIII 258
Query 206 ADASQAMGKVAVQPVSLGV-----DLLTVAGHKVYAPKGVGALYIREGIN----LPPLIL 256
DASQ+ + + D+L +GHK A G G ++I + ++ PL+
Sbjct 259 IDASQSFAHIKYDIKKMKKNKSCPDILITSGHKFCASLGTGFIFINKELSSKYKFKPLLY 318
Query 257 GGG 259
G
Sbjct 319 GSN 321
> tpv:TP01_1094 cysteine desulfurase
Length=469
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 41/137 (29%), Positives = 71/137 (51%), Gaps = 7/137 (5%)
Query 127 EHPAVLRVCEYLRDEEGFELILCPVLSSGLIDLGAFKALLTADT-ILISIPLANGEVGVV 185
EH + L L D G + + +G DL ++LL + + L+ A+ +GV+
Sbjct 182 EHNSNLLPWWVLCDRVGCSIEYVKLHQNGQFDLDHLESLLKSKSPKLLCCGHASNVLGVI 241
Query 186 QPIREVVSVARRI-CPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALY 244
Q ++ + +A + C LV +D++Q +GK+ + + VD L + HK+Y P GVG LY
Sbjct 242 QDMKTISKLAHKYGC---LVLSDSAQTVGKIKIDVQDMDVDFLAGSSHKMYGPTGVGFLY 298
Query 245 IREGI--NLPPLILGGG 259
++ + +L P GGG
Sbjct 299 YKKRLLEDLEPQKCGGG 315
> tgo:TGME49_016170 selenocysteine lyase, putative (EC:2.8.1.7);
K11717 cysteine desulfurase / selenocysteine lyase [EC:2.8.1.7
4.4.1.16]
Length=596
Score = 57.4 bits (137), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 52/193 (26%), Positives = 88/193 (45%), Gaps = 8/193 (4%)
Query 72 LRAAREAAAAALGAP-PCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPA 130
+ R A L A P I+FTSGAT+ IN + + +G IV EH +
Sbjct 170 MEGVRSQLAKFLNADRPEEIVFTSGATDGINLVANTWGEANIGEG---DEIVLTIAEHHS 226
Query 131 VLRVCEYLRDEEGFELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQP-IR 189
L + L + +L + + + + + L+ T L+++ + +G P +
Sbjct 227 NLVPWQLLARRKKAQLKFVELNRDYTLSVSSLVSNLSPRTKLVALSHTSNVLGSFNPYVH 286
Query 190 EVVSVARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVYAPKGVGALYIREGI 249
V + ++I + +V DA+Q++ L D L +GHK+Y P GVG LY + +
Sbjct 287 HVTKLIKQINSNIVVLLDATQSLSHHQTDVRKLKCDFLVGSGHKMYGPTGVGFLYGKYEL 346
Query 250 --NLPPLILGGGQ 260
N+PP GGG+
Sbjct 347 LRNMPPW-KGGGE 358
> mmu:100045796 cysteine desulfurase, mitochondrial-like
Length=148
Score = 50.4 bits (119), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 23/52 (44%), Positives = 34/52 (65%), Gaps = 1/52 (1%)
Query 29 LYLDYNATTPVDPAVFAALSPFLQERFGNPSSK-HAVGVDAAGALRAAREAA 79
LY+D ATTP+DP V A+ P+L +GNP S+ HA G ++ A+ AR+ +
Sbjct 61 LYMDVQATTPLDPRVLDAMLPYLVNYYGNPHSRTHAYGWESEAAMERARQVS 112
> ath:AT5G26600 catalytic/ pyridoxal phosphate binding
Length=475
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Query 184 VVQPIREVVSVARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHK-VYAPKGVGA 242
VV PI+E+V + RR D V DA+ +G V V +G D T HK +AP V
Sbjct 236 VVIPIKELVKICRREGVDQ-VFVDAAHGIGCVDVDMKEIGADFYTSNLHKWFFAPPSVAF 294
Query 243 LYIREGIN 250
LY R+ N
Sbjct 295 LYCRKSSN 302
> mmu:20397 Sgpl1, AI428538, D10Xrf456, S1PL, Spl; sphingosine
phosphate lyase 1 (EC:4.1.2.27); K01634 sphinganine-1-phosphate
aldolase [EC:4.1.2.27]
Length=568
Score = 39.7 bits (91), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 49/195 (25%), Positives = 75/195 (38%), Gaps = 20/195 (10%)
Query 84 GAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEG 143
G P TSG TESI A K+ L KG IV H A + Y G
Sbjct 198 GGPDSCGCVTSGGTESILMACKAYRDLALEKGIKTPEIVAPESAHAAFDKAAHYF----G 253
Query 144 FELILCPVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDAL 203
+++ + + +D+ A K ++ +T ++ GV+ P+ EV +A R
Sbjct 254 MKIVRVALKKNMEVDVQAMKRAISRNTAMLVCSTPQFPHGVMDPVPEVAKLAVRY--KIP 311
Query 204 VHADAS---------QAMGKVAVQPVSL---GVDLLTVAGHKV-YAPKGVG-ALYIREGI 249
+H DA + G +P GV ++ HK YAPKG +Y E
Sbjct 312 LHVDACLGGFLIVFMEKAGYPLEKPFDFRVKGVTSISADTHKYGYAPKGSSVVMYSNEKY 371
Query 250 NLPPLILGGGQESGL 264
+G + G+
Sbjct 372 RTYQFFVGADWQGGV 386
> xla:100037007 sgpl1; sphingosine-1-phosphate lyase 1 (EC:4.1.2.27);
K01634 sphinganine-1-phosphate aldolase [EC:4.1.2.27]
Length=453
Score = 39.7 bits (91), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 47/169 (27%), Positives = 69/169 (40%), Gaps = 19/169 (11%)
Query 84 GAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEG 143
G P TSG TESI A K+ L KG IV H A + Y G
Sbjct 205 GGPNACGTVTSGGTESILMACKAYRDLAYEKGIKHPEIVAPVSAHAAFDKAAHYF----G 260
Query 144 FELILCPVLSSGL-IDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDA 202
+++ PV + + +D+ A K ++ +T ++ G++ PI EV +A +
Sbjct 261 MKIVHIPVNKATMQVDVKAMKRAISKNTAMLVCSAPQFPHGIIDPIEEVAKLALKY--QL 318
Query 203 LVHADASQA------MGKVA--VQPVSL---GVDLLTVAGHKV-YAPKG 239
H DA M K ++P GV ++ HK YAPKG
Sbjct 319 PFHVDACLGGFLIVFMKKAGFPLKPFDFRVKGVTSISADTHKYGYAPKG 367
> hsa:8879 SGPL1, FLJ13811, KIAA1252, SPL; sphingosine-1-phosphate
lyase 1 (EC:4.1.2.27); K01634 sphinganine-1-phosphate aldolase
[EC:4.1.2.27]
Length=568
Score = 37.7 bits (86), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 53/223 (23%), Positives = 80/223 (35%), Gaps = 39/223 (17%)
Query 30 YLDYNATTPVDPAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALGAPPCS 89
Y D+ + P+ P +F L E R A + G P
Sbjct 164 YGDFAWSNPLHPDIFPGLRKIEAE--------------------IVRIACSLFNGGPDSC 203
Query 90 IIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELILC 149
TSG TESI A K+ L KG IV H A + Y G +++
Sbjct 204 GCVTSGGTESILMACKAYRDLAFEKGIKTPEIVAPQSAHAAFNKAASYF----GMKIVRV 259
Query 150 PVLSSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRICPDALVHADAS 209
P+ +D+ A + ++ +T ++ GV+ P+ EV +A + +H DA
Sbjct 260 PLTKMMEVDVRAMRRAISRNTAMLVCSTPQFPHGVIDPVPEVAKLAVKY--KIPLHVDAC 317
Query 210 ---------QAMGKVAVQPVSL---GVDLLTVAGHKV-YAPKG 239
+ G P GV ++ HK YAPKG
Sbjct 318 LGGFLIVFMEKAGYPLEHPFDFRVKGVTSISADTHKYGYAPKG 360
> dre:100037312 sgpl1, zgc:162153; sphingosine-1-phosphate lyase
1 (EC:4.1.2.27); K01634 sphinganine-1-phosphate aldolase
[EC:4.1.2.27]
Length=572
Score = 37.0 bits (84), Expect = 0.085, Method: Compositional matrix adjust.
Identities = 47/179 (26%), Positives = 73/179 (40%), Gaps = 23/179 (12%)
Query 76 REAAAAALGAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVC 135
R A G P TSG TESI A K+ + +G I+ H A +
Sbjct 186 RMTCALFNGGPDSCGTVTSGGTESILMACKAYRDMAHERGIKHPEIIAPISVHAAFDKAA 245
Query 136 EYLRDEEGFELILCPVLSSGL-IDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSV 194
Y G +LI P+ + + +D+ A + +T +T ++ G++ P+ EV +
Sbjct 246 HYF----GMKLIHVPLDNKTMKVDVKAMRRAITKNTAMLVCSAPQFPHGIMDPVEEVAKL 301
Query 195 ARRICPDALVHADA----------SQAMGKVAVQPVSL---GVDLLTVAGHKV-YAPKG 239
A + + H DA +A K+A P GV ++ HK YAPKG
Sbjct 302 AVKY--NIPFHVDACLGGFLIVFMEKAGFKLA--PFDFRVKGVTSISADTHKYGYAPKG 356
> ath:AT1G27980 DPL1; DPL1; carboxy-lyase/ catalytic/ pyridoxal
phosphate binding; K01634 sphinganine-1-phosphate aldolase
[EC:4.1.2.27]
Length=544
Score = 36.2 bits (82), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 40/155 (25%), Positives = 68/155 (43%), Gaps = 9/155 (5%)
Query 42 AVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALGAPPCSIIFTSGATESIN 101
++FA +P + F + + V AL ++E A+ G C + TSG TESI
Sbjct 156 SMFAHTNPLHIDVFQSVVRFESEVVAMTAALLGSKETAS---GGQICGNM-TSGGTESIV 211
Query 102 WALKSGAK-LKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELILCPVLSSGLIDLG 160
A+KS +K KG + ++ H A + +Y + +L PV D+
Sbjct 212 LAVKSSRDYMKYKKGITRPEMIIPESGHSAYDKAAQYFK----IKLWRVPVDKDFRADVK 267
Query 161 AFKALLTADTILISIPLANGEVGVVQPIREVVSVA 195
A + + +TI+I G++ PI E+ +A
Sbjct 268 ATRRHINRNTIMIVGSAPGFPHGIIDPIEELGQLA 302
> tgo:TGME49_012970 protein kinase, putative (EC:2.7.11.17 3.1.3.2)
Length=1196
Score = 34.7 bits (78), Expect = 0.39, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query 125 AMEHPAVLRVCEYLRDEEGFELILCPVLSSGLIDLGAFKALL 166
+ +HPA++++ E++ D+EG LI+ PV G + + + LL
Sbjct 730 SADHPAIIKLVEFIEDDEGVHLIM-PVCKGGPLSRASLRKLL 770
> cel:Y66H1B.4 spl-1; Sphingosine Phosphate Lyase family member
(spl-1); K01634 sphinganine-1-phosphate aldolase [EC:4.1.2.27]
Length=552
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 32/117 (27%), Positives = 46/117 (39%), Gaps = 8/117 (6%)
Query 93 TSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEGFELILCPVL 152
TSG TESI A S G I+ C H A + G L PV
Sbjct 202 TSGGTESIIMACFSYRNRAHSLGIEHPVILACKTAHAAFDKAAHLC----GMRLRHVPVD 257
Query 153 SSGLIDLGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARRI-CPDALVHADA 208
S +DL + L+ ++ ++ N G + PI E+ + ++ P VH DA
Sbjct 258 SDNRVDLKEMERLIDSNVCMLVGSAPNFPSGTIDPIPEIAKLGKKYGIP---VHVDA 311
> cpv:cgd3_520 cysteine-rich extracellular protein with a signal
peptide and two apple domains
Length=683
Score = 31.2 bits (69), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 29/52 (55%), Gaps = 6/52 (11%)
Query 137 YLRDEEGFELILCPVLSSG------LIDLGAFKALLTADTILISIPLANGEV 182
YL + FEL L PVL+ + +G KA LT+++I I+ P + GEV
Sbjct 145 YLGESSYFELTLVPVLNKKGEYVFEIYGVGYEKAHLTSESIGINCPFSIGEV 196
> ath:AT3G62130 epimerase-related
Length=454
Score = 31.2 bits (69), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 2/65 (3%)
Query 184 VVQPIREVVSVARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHK-VYAPKGVGA 242
V+ P+RE+V + R + V DA+ A+G V V +G D HK + P +
Sbjct 209 VLMPVRELVKICREEGVEQ-VFVDAAHAIGSVKVDVKEIGADYYVSNLHKWFFCPPSIAF 267
Query 243 LYIRE 247
Y ++
Sbjct 268 FYCKK 272
> sce:YLR231C BNA5; Bna5p (EC:3.7.1.3); K01556 kynureninase [EC:3.7.1.3]
Length=453
Score = 31.2 bits (69), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 3/67 (4%)
Query 188 IREVVSVARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHKVY--APKGVGALYI 245
I + S A + PD LV D + A+G V +Q GVD +K P G+G L++
Sbjct 208 IGRITSFAHQF-PDILVGWDLAHAVGNVPLQLHDWGVDFACWCSYKYLNAGPGGIGGLFV 266
Query 246 REGINLP 252
P
Sbjct 267 HSKHTKP 273
> tgo:TGME49_100350 molybdopterin cofactor sulfurase, putative
(EC:2.8.1.7)
Length=724
Score = 30.8 bits (68), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 20/76 (26%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Query 29 LYLDYNATTPVD-PAVFAALSPFLQERFGNPSSKHAVGVDAAGALRAAREAAAAALGAP- 86
+Y+DY + + A F +GN S++ L+ AR+ + AP
Sbjct 265 VYMDYAGSGVYQRQQLRAVFDDFTHNAYGNTHSRNPSAKQTDDKLKEARQVISRFFDAPE 324
Query 87 -PCSIIFTSGATESIN 101
++IFTSGAT ++
Sbjct 325 KEYAVIFTSGATAALK 340
> ath:AT1G78070 hypothetical protein
Length=449
Score = 30.8 bits (68), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 29/51 (56%), Gaps = 0/51 (0%)
Query 183 GVVQPIREVVSVARRICPDALVHADASQAMGKVAVQPVSLGVDLLTVAGHK 233
++Q +EV++VA+ I P H SQ++ +V + +++ DL+ G +
Sbjct 161 SLLQRGKEVLNVAKPIVPSMKQHGSLSQSVSRVQISTMAVKDDLIVAGGFQ 211
> cel:Y104H12D.3 hypothetical protein
Length=606
Score = 30.4 bits (67), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 28/117 (23%), Positives = 49/117 (41%), Gaps = 10/117 (8%)
Query 84 GAPPCSIIFTSGATESINWALKSGAKLKRLKGPSPGRIVTCAMEHPAVLRVCEYLRDEEG 143
G C ++ G TE++ A + R +G IV + HPA+ + +
Sbjct 194 GKDSCGVV-AGGGTEALMLACLAYRNRSRARGEWRAEIVAPSTAHPALDKAAAF------ 246
Query 144 FELILCPVLSSGLID---LGAFKALLTADTILISIPLANGEVGVVQPIREVVSVARR 197
F++ + + S D +GA K + T +I N G V PI ++ +A+R
Sbjct 247 FDMTIKRIQVSETDDRANVGAMKRAIGPRTCMIIASAPNHITGTVDPIEKLAKLAQR 303
Lambda K H
0.321 0.138 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10018536124
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40