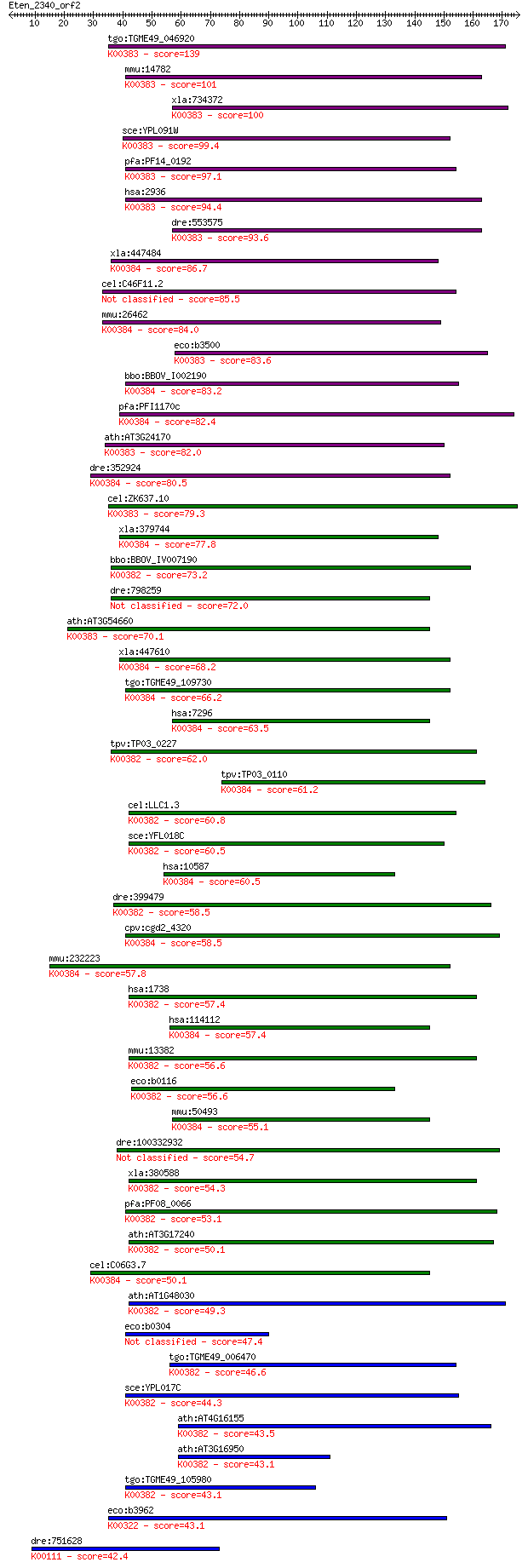
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2340_orf2
Length=175
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7)... 139 5e-33
mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione re... 101 1e-21
xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7); ... 100 3e-21
sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathio... 99.4 5e-21
pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 gluta... 97.1 3e-20
hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7); K0... 94.4 2e-19
dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathi... 93.6 3e-19
xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1... 86.7 4e-17
cel:C46F11.2 hypothetical protein 85.5 7e-17
mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd... 84.0 3e-16
eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreducta... 83.6 3e-16
bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9); ... 83.2 5e-16
pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thiore... 82.4 8e-16
ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase); ... 82.0 1e-15
dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredo... 80.5 2e-15
cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr... 79.3 7e-15
xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1... 77.8 2e-14
bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase (E... 73.2 4e-13
dre:798259 im:7135991; si:ch1073-179p4.3 72.0 9e-13
ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding / gl... 70.1 3e-12
xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1... 68.2 1e-11
tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7)... 66.2 5e-11
hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thior... 63.5 4e-10
tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00... 62.0 9e-10
tpv:TP03_0110 thioredoxin reductase (EC:1.6.4.5); K00384 thior... 61.2 2e-09
cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehyd... 60.8 2e-09
sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the li... 60.5 3e-09
hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin r... 60.5 3e-09
dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase (E... 58.5 1e-08
cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin red... 58.5 1e-08
mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase 3... 57.8 2e-08
hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehy... 57.4 2e-08
hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3 (E... 57.4 2e-08
mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogen... 56.6 4e-08
eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydroge... 56.6 4e-08
mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:... 55.1 1e-07
dre:100332932 glutathione reductase-like 54.7 2e-07
xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1... 54.3 2e-07
pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4); ... 53.1 5e-07
ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP ... 50.1 4e-06
cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-... 50.1 4e-06
ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrog... 49.3 6e-06
eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide... 47.4 3e-05
tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.... 46.6 5e-05
sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogena... 44.3 2e-04
ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoa... 43.5 4e-04
ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrol... 43.1 4e-04
tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putativ... 43.1 4e-04
eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotid... 43.1 5e-04
dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe0... 42.4 9e-04
> tgo:TGME49_046920 glutathione reductase, putative (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 139 bits (350), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 70/141 (49%), Positives = 93/141 (65%), Gaps = 5/141 (3%)
Query 35 MAAA--HHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCA 92
MAAA HFD V+GGGSGG+A ARRAATY +V L + +R+GGTCVNVGCVPKK+MWC
Sbjct 5 MAAATRRHFDLFVIGGGSGGLACARRAATYNVRVGLADGNRLGGTCVNVGCVPKKVMWCV 64
Query 93 ANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFAT 152
A+ E+L +K+ + P W+ L NR+NYIKRLN IY +NL S V + +A
Sbjct 65 ASVHETLHELKNFAFTVKEQPTFCWRTLKTNRDNYIKRLNNIYLNNLKNSGVTFFPAYAR 124
Query 153 L-KPKDEANG--QHVVVVNSS 170
KP+ + +G H +V+ S+
Sbjct 125 FAKPEAKTDGGLAHAIVLKSA 145
> mmu:14782 Gsr, AI325518, D8Ertd238e, Gr-1, Gr1; glutathione
reductase (EC:1.8.1.7); K00383 glutathione reductase (NADPH)
[EC:1.8.1.7]
Length=500
Score = 101 bits (252), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 59/126 (46%), Positives = 81/126 (64%), Gaps = 5/126 (3%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
FD +V+GGGSGG+A ARRAA GA+ A+VE ++GGTCVNVGCVPKK+MW A E +
Sbjct 43 FDYLVIGGGSGGLASARRAAELGARAAVVESHKLGGTCVNVGCVPKKVMWNTAVHSEFMH 102
Query 101 GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL----KPK 156
G + S + +W + Q R+ Y+ RLN IY++NL KS + +G+AT +P
Sbjct 103 DHVDYGFQ-SCEGKFSWHVIKQKRDAYVSRLNTIYQNNLTKSHIEIIHGYATFADGPRPT 161
Query 157 DEANGQ 162
E NG+
Sbjct 162 VEVNGK 167
> xla:734372 gsr, MGC84926; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=476
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 53/124 (42%), Positives = 72/124 (58%), Gaps = 10/124 (8%)
Query 57 RRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLN 116
RRAA GA+ A+VE S++GGTCVNVGCVPKKIMW AA E + G E D +
Sbjct 35 RRAAELGARTAVVESSKLGGTCVNVGCVPKKIMWNAAMHSEYIHDHADYGFEIPD-VKFT 93
Query 117 WQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL----KPKDEANGQ-----HVVVV 167
W+ + + R+ Y+ RLN IY++NL K+Q+ G A +P E NGQ H+++
Sbjct 94 WKVIKEKRDAYVSRLNDIYQNNLQKAQIEIIRGNANFTSDPEPTVEVNGQKYSAPHILIA 153
Query 168 NSSK 171
K
Sbjct 154 TGGK 157
> sce:YPL091W GLR1, LPG17; Cytosolic and mitochondrial glutathione
oxidoreductase, converts oxidized glutathione to reduced
glutathione; mitochondrial but not cytosolic form has a role
in resistance to hyperoxia (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=483
Score = 99.4 bits (246), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 56/118 (47%), Positives = 76/118 (64%), Gaps = 7/118 (5%)
Query 40 HFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESL 99
H+D +V+GGGSGG+A ARRAA+YGAK LVE +GGTCVNVGCVPKK+MW A++ +
Sbjct 23 HYDYLVIGGGSGGVASARRAASYGAKTLLVEAKALGGTCVNVGCVPKKVMWYASDLATRV 82
Query 100 QGMKHLGVEFSDPP------RLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFA 151
G+ + + P NW Q R+ Y+ RLN IY+ NL+K +V +G+A
Sbjct 83 SHANEYGL-YQNLPLDKEHLTFNWPEFKQKRDAYVHRLNGIYQKNLEKEKVDVVFGWA 139
> pfa:PF14_0192 glutathione reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=500
Score = 97.1 bits (240), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 62/113 (54%), Positives = 79/113 (69%), Gaps = 2/113 (1%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
+D +V+GGGSGG+A ARRAA + AKVALVE+SR+GGTCVNVGCVPKKIM+ AA+ + L+
Sbjct 3 YDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDILE 62
Query 101 GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
+H G F N LV+ R+ YI+RLN IY NL K +V Y G A+
Sbjct 63 NSRHYG--FDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASF 113
> hsa:2936 GSR, MGC78522; glutathione reductase (EC:1.8.1.7);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=522
Score = 94.4 bits (233), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 57/126 (45%), Positives = 78/126 (61%), Gaps = 5/126 (3%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
+D +V+GGGSGG+A ARRAA GA+ A+VE ++GGTCVNVGCVPKK+MW A E +
Sbjct 65 YDYLVIGGGSGGLASARRAAELGARAAVVESHKLGGTCVNVGCVPKKVMWNTAVHSEFMH 124
Query 101 GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL----KPK 156
G S + NW+ + + R+ Y+ RLN IY++NL KS + G A KP
Sbjct 125 DHADYGFP-SCEGKFNWRVIKEKRDAYVSRLNAIYQNNLTKSHIEIIRGHAAFTSDPKPT 183
Query 157 DEANGQ 162
E +G+
Sbjct 184 IEVSGK 189
> dre:553575 MGC110010; zgc:110010 (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=425
Score = 93.6 bits (231), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 46/110 (41%), Positives = 65/110 (59%), Gaps = 5/110 (4%)
Query 57 RRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLN 116
RRAA GA A++E R+GGTCVNVGCVPKK+MW + E L + G E + +
Sbjct 25 RRAAELGATTAVIESHRLGGTCVNVGCVPKKVMWNTSTHAEYLHDHEDYGFEGA-KAHFS 83
Query 117 WQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL----KPKDEANGQ 162
WQ + R+ Y+ RLN+IY NL+K ++ +G+A +P E NG+
Sbjct 84 WQIIKHKRDAYVSRLNQIYRSNLEKGKIEFIHGYARFTDDPEPTVEVNGK 133
> xla:447484 txnrd3, MGC81848; thioredoxin reductase 3 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=596
Score = 86.7 bits (213), Expect = 4e-17, Method: Composition-based stats.
Identities = 46/121 (38%), Positives = 72/121 (59%), Gaps = 10/121 (8%)
Query 36 AAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPK 86
+ + +D +V+GGGSGG+A ++ AA++G KV +++ +GGTCVNVGC+PK
Sbjct 107 SVTYDYDLIVIGGGSGGLACSKEAASFGKKVMVLDFVVPTPQGTSWGLGGTCVNVGCIPK 166
Query 87 KIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHR 146
K+M AA +SL+ + G E+ + + NW+ + + +NYI LN Y L QV R
Sbjct 167 KLMHQAAILGQSLKDSRKFGWEYEEQVKHNWETMREAIQNYIGSLNWGYRVALRDKQV-R 225
Query 147 Y 147
Y
Sbjct 226 Y 226
> cel:C46F11.2 hypothetical protein
Length=473
Score = 85.5 bits (210), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 56/122 (45%), Positives = 78/122 (63%), Gaps = 3/122 (2%)
Query 33 SAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMW-C 91
S M+ FD +V+GGGSGGIA ARRA +G V L+E R+GGTCVNVGCVPKK+M+ C
Sbjct 13 SIMSGVKEFDYLVIGGGSGGIASARRAREFGVSVGLIESGRLGGTCVNVGCVPKKVMYNC 72
Query 92 AANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFA 151
+ +A E ++ G + + + +W+ + ++R+ YIKRLN +YE L S V G A
Sbjct 73 SLHA-EFIRDHADYGFDVT-LNKFDWKVIKKSRDEYIKRLNGLYESGLKGSSVEYIRGRA 130
Query 152 TL 153
T
Sbjct 131 TF 132
> mmu:26462 Txnrd2, AA118373, ESTM573010, TGR, Tr3, Trxr2, Trxrd2;
thioredoxin reductase 2 (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=527
Score = 84.0 bits (206), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 72/125 (57%), Gaps = 10/125 (8%)
Query 33 SAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVAL---VERSR------MGGTCVNVGC 83
SA FD +V+GGGSGG+A A+ AA G KVA+ VE S +GGTCVNVGC
Sbjct 35 SAAGGQQSFDLLVIGGGSGGLACAKEAAQLGKKVAVADYVEPSPRGTKWGLGGTCVNVGC 94
Query 84 VPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQ 143
+PKK+M AA ++ H G E + P + NW+ + + +N++K LN + L +
Sbjct 95 IPKKLMHQAALLGGMIRDAHHYGWEVAQPVQHNWKTMAEAVQNHVKSLNWGHRVQLQDRK 154
Query 144 VHRYY 148
V +Y+
Sbjct 155 V-KYF 158
> eco:b3500 gor, ECK3485, gorA, JW3467; glutathione oxidoreductase
(EC:1.8.1.7); K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=450
Score = 83.6 bits (205), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 46/112 (41%), Positives = 64/112 (57%), Gaps = 8/112 (7%)
Query 58 RAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEF---SDPPR 114
RAA YG K AL+E +GGTCVNVGCVPKK+MW AA E++ G ++ + +
Sbjct 22 RAAMYGQKCALIEAKELGGTCVNVGCVPKKVMWHAAQIREAIH---MYGPDYGFDTTINK 78
Query 115 LNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKD--EANGQHV 164
NW+ L+ +R YI R++ YE+ L K+ V GFA E NG+ +
Sbjct 79 FNWETLIASRTAYIDRIHTSYENVLGKNNVDVIKGFARFVDAKTLEVNGETI 130
> bbo:BBOV_I002190 19.m02214; thiodoxin reductase (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=559
Score = 83.2 bits (204), Expect = 5e-16, Method: Composition-based stats.
Identities = 48/124 (38%), Positives = 68/124 (54%), Gaps = 12/124 (9%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVPKKIMWC 91
+D V+GGG G+A A+ AA GAK L + R +GGTCVNVGC+PKK+M
Sbjct 69 YDLAVIGGGCSGLAAAKEAARLGAKTVLFDYVRPSPRGTKWGLGGTCVNVGCIPKKLMHY 128
Query 92 AANAFESLQGMKHLGVEFSD-PPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF 150
A + + LG +SD P+ +W +++Q +NY+K LN Y L + V F
Sbjct 129 AGILGHAEHDREMLG--WSDASPKHDWSKMIQTIQNYVKMLNFSYRSGLLTTGVKYINAF 186
Query 151 ATLK 154
ATL+
Sbjct 187 ATLE 190
> pfa:PFI1170c thioredoxin reductase (EC:1.8.1.9); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=617
Score = 82.4 bits (202), Expect = 8e-16, Method: Composition-based stats.
Identities = 54/145 (37%), Positives = 82/145 (56%), Gaps = 13/145 (8%)
Query 39 HHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVPKKIM 89
+ +D VV+GGG GG+A A+ AA +GA+V L + + +GGTCVNVGCVPKK+M
Sbjct 116 YDYDYVVIGGGPGGMASAKEAAAHGARVLLFDYVKPSSQGTKWGIGGTCVNVGCVPKKLM 175
Query 90 WCAANAFESLQ-GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYY 148
A + + K G +F D + +W++LV +++I+ LN Y L S+V
Sbjct 176 HYAGHMGSIFKLDSKAYGWKF-DNLKHDWKKLVTTVQSHIRSLNFSYMTGLRSSKVKYIN 234
Query 149 GFATLKPKDEANGQHVVVVNSSKEE 173
G A LK K+ + + + + SKEE
Sbjct 235 GLAKLKDKNTVS--YYLKGDLSKEE 257
> ath:AT3G24170 ATGR1; ATGR1 (glutathione-disulfide reductase);
FAD binding / NADP or NADPH binding / glutathione-disulfide
reductase/ oxidoreductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=499
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 67/126 (53%), Gaps = 10/126 (7%)
Query 34 AMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER----------SRMGGTCVNVGC 83
A A + FD V+G GSGG+ AR +A +GAKV + E +GGTCV GC
Sbjct 19 ANATHYDFDLFVIGAGSGGVRAARFSANHGAKVGICELPFHPISSEEIGGVGGTCVIRGC 78
Query 84 VPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQ 143
VPKKI+ A L+ K+ G E ++ W++L+Q + + I RLN IY+ L +
Sbjct 79 VPKKILVYGATYGGELEDAKNYGWEINEKVDFTWKKLLQKKTDEILRLNNIYKRLLANAA 138
Query 144 VHRYYG 149
V Y G
Sbjct 139 VKLYEG 144
> dre:352924 txnrd1, TrxR1, cb682, fb83a08, wu:fb83a08; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=602
Score = 80.5 bits (197), Expect = 2e-15, Method: Composition-based stats.
Identities = 45/132 (34%), Positives = 70/132 (53%), Gaps = 9/132 (6%)
Query 29 QPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERS---------RMGGTCV 79
Q L + + +D +V+GGGSGG+A ++ AAT G KV +++ +GGTCV
Sbjct 104 QKLLGEGSEVYDYDLIVIGGGSGGLACSKEAATLGKKVMVLDYVVPTPQGTAWGLGGTCV 163
Query 80 NVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNL 139
NVGC+PKK+M A +++ + G EF++ NW+ + NYI LN Y +L
Sbjct 164 NVGCIPKKLMHQTALLGTAMEDARKFGWEFAEQVTHNWETMKTAVNNYIGSLNWGYRVSL 223
Query 140 DKSQVHRYYGFA 151
V+ +A
Sbjct 224 RDKNVNYVNAYA 235
> cel:ZK637.10 trxr-2; ThioRedoXin Reductase family member (trxr-2);
K00383 glutathione reductase (NADPH) [EC:1.8.1.7]
Length=503
Score = 79.3 bits (194), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 46/151 (30%), Positives = 81/151 (53%), Gaps = 16/151 (10%)
Query 35 MAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVP 85
+ +++ FD +V+G GSGG++ ++RAA GA VAL++ +GGTC NVGC+P
Sbjct 15 LFSSNKFDLIVIGAGSGGLSCSKRAADLGANVALIDAVEPTPHGHSWGIGGTCANVGCIP 74
Query 86 KKIMWCAANAFESLQGMKHLGVEFSDPPRL--NWQRLVQNRENYIKRLNRIYEDNLDKSQ 143
KK+M AA + L+ G D ++ +W L +N + +K N IY L++ +
Sbjct 75 KKLMHQAAIVGKELKHADKYGWNGIDQEKIKHDWNVLSKNVNDRVKANNWIYRVQLNQKK 134
Query 144 VHRYYGFATLKPKDEANGQHVVVVNSSKEEA 174
++ + +A KD+ +V+ + K +
Sbjct 135 INYFNAYAEFVDKDK-----IVITGTDKNKT 160
> xla:379744 txnrd2, MGC69182; thioredoxin reductase 2 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=504
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/118 (38%), Positives = 68/118 (57%), Gaps = 9/118 (7%)
Query 39 HHFDCVVLGGGSGGIAFARRAATYGAKVAL---VERSR------MGGTCVNVGCVPKKIM 89
H +D +V+GGGSGG+A A++AA +G KVA+ VE S +GGTCVNVGC+PKK+M
Sbjct 18 HDYDLLVIGGGSGGLACAKQAAQFGKKVAVFDYVEPSPRGTKWGIGGTCVNVGCIPKKLM 77
Query 90 WCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRY 147
AA +++ H G + +W ++ +NY+K LN + L +V +
Sbjct 78 HQAALIGSTMKDAPHYGWGTPYEIQHDWGKMAGAVQNYVKSLNWGHRIQLQDKKVKYF 135
> bbo:BBOV_IV007190 23.m05858; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=481
Score = 73.2 bits (178), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 69/128 (53%), Gaps = 11/128 (8%)
Query 36 AAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAAN 94
+A H +D V+GGG GG A +AA YG KVA ++ R+ +GGTC+NVGC+P K + ++
Sbjct 19 SATHKYDLAVIGGGPGGYTTAIKAAQYGLKVACIDRRTTLGGTCLNVGCIPSKCLLNTSH 78
Query 95 AF----ESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF 150
+ + + G+K VEF N + + ++ +K L+ + K+ V G
Sbjct 79 HYKASHDGIAGIKFTNVEF------NHGQTMSSKAKILKTLDAGIKGLFKKNGVDYISGH 132
Query 151 ATLKPKDE 158
TLK +E
Sbjct 133 GTLKSANE 140
> dre:798259 im:7135991; si:ch1073-179p4.3
Length=371
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 39/118 (33%), Positives = 66/118 (55%), Gaps = 9/118 (7%)
Query 36 AAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPK 86
A +D VV+GGGSGG+A ++ AA G KVA+++ + +GGTCVNVGC+PK
Sbjct 28 AGKFDYDLVVIGGGSGGLACSKEAAQLGQKVAVLDYVEPSLKGTKWGLGGTCVNVGCIPK 87
Query 87 KIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQV 144
K+M AA +++ + G + + +W + + +N+++ LN + L +V
Sbjct 88 KLMHQAALLGTAVKDARKYGWQIPETLSHDWPTMAEAVQNHVRSLNWGHRVQLQDKKV 145
> ath:AT3G54660 GR; GR (GLUTATHIONE REDUCTASE); ATP binding /
glutathione-disulfide reductase (EC:1.8.1.7); K00383 glutathione
reductase (NADPH) [EC:1.8.1.7]
Length=565
Score = 70.1 bits (170), Expect = 3e-12, Method: Composition-based stats.
Identities = 41/134 (30%), Positives = 66/134 (49%), Gaps = 10/134 (7%)
Query 21 QRDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE---------- 70
+R S AS + + + FD +G GSGG+ +R A ++GA A+ E
Sbjct 68 RRFSVCASTDNGAESDRHYDFDLFTIGAGSGGVRASRFATSFGASAAVCELPFSTISSDT 127
Query 71 RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKR 130
+GGTCV GCVPKK++ A+ + G ++ P +W L+ N+ ++R
Sbjct 128 AGGVGGTCVLRGCVPKKLLVYASKYSHEFEDSHGFGWKYETEPSHDWTTLIANKNAELQR 187
Query 131 LNRIYEDNLDKSQV 144
L IY++ L K+ V
Sbjct 188 LTGIYKNILSKANV 201
> xla:447610 txnrd1, MGC85342; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=531
Score = 68.2 bits (165), Expect = 1e-11, Method: Composition-based stats.
Identities = 44/122 (36%), Positives = 72/122 (59%), Gaps = 9/122 (7%)
Query 39 HHFDCVVLGGGSGGIAFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIM 89
+ +D +V+GGGSGG+A ++ AA YG KV +++ + +GGTCVNVGC+PKK+M
Sbjct 46 YDYDLIVIGGGSGGLAASKEAAKYGKKVLVLDFVTPSPLGTKWGLGGTCVNVGCIPKKLM 105
Query 90 WCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYG 149
AA ++L+ + G + +D + NW+ + + +NYI LN Y L ++ V G
Sbjct 106 HQAALLRQALKDSQKYGWQIADNIQHNWETMTDSVQNYIGSLNFNYRVALMENNVKYENG 165
Query 150 FA 151
+
Sbjct 166 YG 167
> tgo:TGME49_109730 thioredoxin reductase, putative (EC:1.8.1.7);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=662
Score = 66.2 bits (160), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 63/124 (50%), Gaps = 16/124 (12%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVPKKIM-- 89
+D V+GGGSGG+A A+ AA GA+ + + + +GGTCVNVGCVPK +
Sbjct 160 YDLAVIGGGSGGLACAKMAAAQGAETVVFDFVQPSTQGSTWGLGGTCVNVGCVPKYLFHH 219
Query 90 --WCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRY 147
ANA H+G + +++W V+ +NYIK LN Y L K+ V
Sbjct 220 TGLAGANAH---WDGPHMGWKGKFEEQVDWGVCVEKVQNYIKSLNFGYRTGLRKAGVTYI 276
Query 148 YGFA 151
+A
Sbjct 277 NAYA 280
> hsa:7296 TXNRD1, GRIM-12, MGC9145, TR, TR1, TRXR1, TXNR; thioredoxin
reductase 1 (EC:1.8.1.9); K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=649
Score = 63.5 bits (153), Expect = 4e-10, Method: Composition-based stats.
Identities = 34/97 (35%), Positives = 56/97 (57%), Gaps = 9/97 (9%)
Query 57 RRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGV 107
+ AA YG KV +++ R +GGTCVNVGC+PKK+M AA ++LQ ++ G
Sbjct 179 KEAAQYGKKVMVLDFVTPTPLGTRWGLGGTCVNVGCIPKKLMHQAALLGQALQDSRNYGW 238
Query 108 EFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQV 144
+ + + +W R+++ +N+I LN Y L + +V
Sbjct 239 KVEETVKHDWDRMIEAVQNHIGSLNWGYRVALREKKV 275
> tpv:TP03_0227 dihydrolipoamide dehydrogenase (EC:1.8.1.4); K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 69/130 (53%), Gaps = 11/130 (8%)
Query 36 AAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAAN 94
++ +D +VLG G GG A +AA +G KV +VE R +GGTC+N GC+P K + ++
Sbjct 19 TSSSKYDLLVLGAGPGGYTMAIKAAQHGLKVGVVEKRPTLGGTCLNCGCIPSKSLLNTSH 78
Query 95 AF----ESLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF 150
+ + + G++ G+E + ++++ +++ ++ LN K+++ G
Sbjct 79 LYHLMKKGVNGLRITGLE------TDVGKMMEEKDSVMRTLNMGIFGLFKKNKIDYIQGT 132
Query 151 ATLKPKDEAN 160
A K ++E
Sbjct 133 ACFKSQNEVT 142
> tpv:TP03_0110 thioredoxin reductase (EC:1.6.4.5); K00384 thioredoxin
reductase (NADPH) [EC:1.8.1.9]
Length=567
Score = 61.2 bits (147), Expect = 2e-09, Method: Composition-based stats.
Identities = 33/91 (36%), Positives = 47/91 (51%), Gaps = 1/91 (1%)
Query 74 MGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFS-DPPRLNWQRLVQNRENYIKRLN 132
+GGTCVNVGC+PKK+M A+ + G+ S D ++NW +L Q +NY+K LN
Sbjct 133 VGGTCVNVGCIPKKLMHYASLLRSTQYDRFQYGITSSPDQEQVNWTKLTQTIQNYVKMLN 192
Query 133 RIYEDNLDKSQVHRYYGFATLKPKDEANGQH 163
Y L + V + TLK + H
Sbjct 193 FSYRSGLTTAGVDYINAYGTLKHNNTVEYTH 223
> cel:LLC1.3 hypothetical protein; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=495
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 37/114 (32%), Positives = 61/114 (53%), Gaps = 4/114 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVER-SRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
D VV+GGG GG A +AA G K VE+ + +GGTC+NVGC+P K + ++ Q
Sbjct 31 DLVVIGGGPGGYVAAIKAAQLGMKTVCVEKNATLGGTCLNVGCIPSKALLNNSHYLHMAQ 90
Query 101 -GMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
G++ + LN ++++ + N +K+L + ++V GFAT+
Sbjct 91 HDFAARGIDCT--ASLNLPKMMEAKSNSVKQLTGGIKQLFKANKVGHVEGFATI 142
> sce:YFL018C LPD1, HPD1; Dihydrolipoamide dehydrogenase, the
lipoamide dehydrogenase component (E3) of the pyruvate dehydrogenase
and 2-oxoglutarate dehydrogenase multi-enzyme complexes
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=499
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 34/110 (30%), Positives = 59/110 (53%), Gaps = 2/110 (1%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V++GGG G A +AA G A VE R ++GGTC+NVGC+P K + ++ F +
Sbjct 28 DVVIIGGGPAGYVAAIKAAQLGFNTACVEKRGKLGGTCLNVGCIPSKALLNNSHLFHQMH 87
Query 101 G-MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYG 149
+ G++ + ++N + +++ +K+L E K++V Y G
Sbjct 88 TEAQKRGIDVNGDIKINVANFQKAKDDAVKQLTGGIELLFKKNKVTYYKG 137
> hsa:10587 TXNRD2, SELZ, TR, TR-BETA, TR3, TRXR2; thioredoxin
reductase 2 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH)
[EC:1.8.1.9]
Length=524
Score = 60.5 bits (145), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 34/88 (38%), Positives = 51/88 (57%), Gaps = 9/88 (10%)
Query 54 AFARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKH 104
A A+ AA G KVA+V+ R +GGTCVNVGC+PKK+M AA +Q +
Sbjct 53 ACAKEAAQLGRKVAVVDYVEPSPQGTRWGLGGTCVNVGCIPKKLMHQAALLGGLIQDAPN 112
Query 105 LGVEFSDPPRLNWQRLVQNRENYIKRLN 132
G E + P +W+++ + +N++K LN
Sbjct 113 YGWEVAQPVPHDWRKMAEAVQNHVKSLN 140
> dre:399479 dldh, wu:fb24b05; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/137 (29%), Positives = 67/137 (48%), Gaps = 9/137 (6%)
Query 37 AAHHFDCVVLGGGSGGIAFARRAATYGAKVALVER-SRMGGTCVNVGCVPKKIMWCAANA 95
AA D V+G G GG A +AA G K VE+ + +GGTC+NVGC+P K + +
Sbjct 37 AAIDADVTVVGSGPGGYVAAIKAAQLGFKTVCVEKNATLGGTCLNVGCIPSKALLNNSYL 96
Query 96 FESLQG--MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
+ G + G+E LN ++++ + +K L +++V GF T+
Sbjct 97 YHMAHGKDFESRGIEIQG-ISLNLEKMMAQKSGAVKALTGGIAHLFKQNKVTHVNGFGTI 155
Query 154 KPKDE-----ANGQHVV 165
K++ A+G+ V+
Sbjct 156 TGKNQVTAKTADGEQVI 172
> cpv:cgd2_4320 thioredoxin reductase 1 ; K00384 thioredoxin reductase
(NADPH) [EC:1.8.1.9]
Length=526
Score = 58.5 bits (140), Expect = 1e-08, Method: Composition-based stats.
Identities = 54/142 (38%), Positives = 72/142 (50%), Gaps = 16/142 (11%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCVNVGCVPKKIMWC 91
+D VV+GGGSGG+A A+ AA YG KVAL + + +GGTCVNVGCVPKK+M
Sbjct 33 YDLVVIGGGSGGMAAAKEAAKYGKKVALFDFVKPSTQGTKWGLGGTCVNVGCVPKKLMHY 92
Query 92 AANAFESL-QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF 150
+A S+ + G + S W +LV+ N+I+ LN Y L V
Sbjct 93 SALIASSIHHDAQMFGHKTSS--SFEWGKLVETLRNHIRMLNFSYRTGLRVGNVEYINAL 150
Query 151 ATL-KPKD---EANGQHVVVVN 168
A L P E NGQ + +
Sbjct 151 AKLIDPHSVEYEDNGQKKTITS 172
> mmu:232223 Txnrd3, AI196535, TR2, Tgr; thioredoxin reductase
3 (EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=652
Score = 57.8 bits (138), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 50/146 (34%), Positives = 75/146 (51%), Gaps = 22/146 (15%)
Query 15 LQQQLQQRDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVE---- 70
L Q+L Q DSA H +D +++GGGSGG++ A+ AA G KV +++
Sbjct 153 LLQKLLQDDSA-------------HDYDLIIIGGGSGGLSCAKEAANLGKKVMVLDFVVP 199
Query 71 -----RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPPRLNWQRLVQNRE 125
+GGTCVNVGC+PKK+M AA +LQ K G E++ + NW+ + + +
Sbjct 200 SPQGTTWGLGGTCVNVGCIPKKLMHQAALLGHALQDAKKYGWEYNQQVKHNWEAMTEAIQ 259
Query 126 NYIKRLNRIYEDNLDKSQVHRYYGFA 151
++I LN Y L + V F
Sbjct 260 SHIGSLNWGYRVTLREKGVTYVNSFG 285
> hsa:1738 DLD, DLDH, E3, GCSL, LAD, PHE3; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=509
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 61/122 (50%), Gaps = 4/122 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V+G G GG A +AA G K +E++ +GGTC+NVGC+P K + ++ +
Sbjct 43 DVTVIGSGPGGYVAAIKAAQLGFKTVCIEKNETLGGTCLNVGCIPSKALLNNSHYYHMAH 102
Query 101 G--MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDE 158
G G+E S+ RLN ++++ + +K L +++V G+ + K++
Sbjct 103 GKDFASRGIEMSE-VRLNLDKMMEQKSTAVKALTGGIAHLFKQNKVVHVNGYGKITGKNQ 161
Query 159 AN 160
Sbjct 162 VT 163
> hsa:114112 TXNRD3, TGR, TR2, TRXR3; thioredoxin reductase 3
(EC:1.8.1.9); K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=607
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 53/98 (54%), Gaps = 9/98 (9%)
Query 56 ARRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLG 106
A+ AA G KV +++ +GGTCVNVGC+PKK+M AA ++L + G
Sbjct 172 AKEAAILGKKVMVLDFVVPSPQGTSWGLGGTCVNVGCIPKKLMHQAALLGQALCDSRKFG 231
Query 107 VEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQV 144
E++ R NW+ + + +N+I LN Y +L + V
Sbjct 232 WEYNQQVRHNWETMTKAIQNHISSLNWGYRLSLREKAV 269
> mmu:13382 Dld, AI315664, AI746344; dihydrolipoamide dehydrogenase
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 62/122 (50%), Gaps = 4/122 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V+G G GG A ++A G K +E++ +GGTC+NVGC+P K + ++ +
Sbjct 43 DVTVIGSGPGGYVAAIKSAQLGFKTVCIEKNETLGGTCLNVGCIPSKALLNNSHYYHMAH 102
Query 101 G--MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDE 158
G G+E + RLN +++++ + + +K L +++V GF + K++
Sbjct 103 GKDFASRGIEIPE-VRLNLEKMMEQKHSAVKALTGGIAHLFKQNKVVHVNGFGKITGKNQ 161
Query 159 AN 160
Sbjct 162 VT 163
> eco:b0116 lpd, dhl, ECK0115, JW0112, lpdA; lipoamide dehydrogenase,
E3 component is part of three enzyme complexes (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=474
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 48/91 (52%), Gaps = 2/91 (2%)
Query 43 CVVLGGGSGGIAFARRAATYGAKVALVER-SRMGGTCVNVGCVPKKIMWCAANAFESLQG 101
VVLG G G + A R A G + +VER + +GG C+NVGC+P K + A E +
Sbjct 9 VVVLGAGPAGYSAAFRCADLGLETVIVERYNTLGGVCLNVGCIPSKALLHVAKVIEEAKA 68
Query 102 MKHLGVEFSDPPRLNWQRLVQNRENYIKRLN 132
+ G+ F + P+ + ++ +E I +L
Sbjct 69 LAEHGIVFGE-PKTDIDKIRTWKEKVINQLT 98
> mmu:50493 Txnrd1, TR, TR1, TrxR1; thioredoxin reductase 1 (EC:1.8.1.9);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=499
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 30/97 (30%), Positives = 56/97 (57%), Gaps = 9/97 (9%)
Query 57 RRAATYGAKVALVE---------RSRMGGTCVNVGCVPKKIMWCAANAFESLQGMKHLGV 107
+ AA + KV +++ R +GGTCVNVGC+PKK+M AA ++L+ ++ G
Sbjct 29 KEAAKFDKKVLVLDFVTPTPLGTRWGLGGTCVNVGCIPKKLMHQAALLGQALKDSRNYGW 88
Query 108 EFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQV 144
+ D + +W+++ ++ +++I LN Y L + +V
Sbjct 89 KVEDTVKHDWEKMTESVQSHIGSLNWGYRVALREKKV 125
> dre:100332932 glutathione reductase-like
Length=461
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/140 (30%), Positives = 67/140 (47%), Gaps = 10/140 (7%)
Query 38 AHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFE 97
++ +D V+GGGSGG+ R AA+ G KV + E R GGTCV GCVPKK+ A+ E
Sbjct 3 SYDYDLFVIGGGSGGVRSGRVAASMGKKVGIAEEYRYGGTCVIRGCVPKKLFVYASQFHE 62
Query 98 SLQGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL---- 153
+ G + +W +L+ ++ I RL +Y L+ ++ + A L
Sbjct 63 HFEDSAGFGWTVGETS-FDWAKLIAAKDREIDRLEGLYRKGLENAKAKVFDSRAELVDAH 121
Query 154 -----KPKDEANGQHVVVVN 168
K + +H+V+
Sbjct 122 TVRLTKTGQTVSAEHIVIAT 141
> xla:380588 dld, MGC68940; dihydrolipoamide dehydrogenase (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=509
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/122 (27%), Positives = 60/122 (49%), Gaps = 4/122 (3%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVERS-RMGGTCVNVGCVPKKIMWCAANAFESLQ 100
D V+G G GG A +AA G + VE++ +GGTC+NVGC+P K + ++ +
Sbjct 43 DVTVVGSGPGGYVAAIKAAQLGFQTVCVEKNDTLGGTCLNVGCIPSKALLNNSHLYHLAH 102
Query 101 G--MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDE 158
G G+E + LN +++++ + +K L +++V GF + K++
Sbjct 103 GKDFASRGIEVTG-IHLNLEKMMEQKSGAVKSLTSGIAHLFKQNKVVHVQGFGKITGKNQ 161
Query 159 AN 160
Sbjct 162 VT 163
> pfa:PF08_0066 lipoamide dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=666
Score = 53.1 bits (126), Expect = 5e-07, Method: Composition-based stats.
Identities = 42/138 (30%), Positives = 67/138 (48%), Gaps = 13/138 (9%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALV--ERSRMGGTCVNVGCVPKKIMWCAANAFES 98
+D ++G G GG A A A KV + + + +GGTCVNVGC+P K + A N +
Sbjct 126 YDLAIIGCGVGGHAAAINAMERNLKVIIFAGDENCIGGTCVNVGCIPSKALLYATNKYRE 185
Query 99 LQGMKHL---GV-----EFSDPPRLNWQRLVQNR-ENYIKRLNRIYEDNLDKSQVHRYYG 149
L+ + L G+ + + + +LV N + I +L + +DK + +G
Sbjct 186 LKNLDKLYYYGIHSNIFQNNKNTEIENNQLVSNSFQINITKLKEYTQSVIDKLRNGISHG 245
Query 150 FATLKPKDEANGQHVVVV 167
F TL K N +HV V+
Sbjct 246 FKTL--KFNKNSEHVQVI 261
> ath:AT3G17240 mtLPD2; mtLPD2 (LIPOAMIDE DEHYDROGENASE 2); ATP
binding / dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=507
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 42/132 (31%), Positives = 71/132 (53%), Gaps = 8/132 (6%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAF-ESL 99
D V++GGG GG A +AA G K +E R +GGTC+NVGC+P K + +++ + E+
Sbjct 45 DVVIIGGGPGGYVAAIKAAQLGLKTTCIEKRGALGGTCLNVGCIPSKALLHSSHMYHEAK 104
Query 100 QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFAT-LKPK-- 156
+ GV+ S ++ ++ ++ +K L R E K++V+ G+ L P
Sbjct 105 HVFANHGVKVSS-VEVDLPAMLAQKDTAVKNLTRGVEGLFKKNKVNYVKGYGKFLSPSEV 163
Query 157 --DEANGQHVVV 166
D +G++VVV
Sbjct 164 SVDTIDGENVVV 175
> cel:C06G3.7 trxr-1; ThioRedoXin Reductase family member (trxr-1);
K00384 thioredoxin reductase (NADPH) [EC:1.8.1.9]
Length=667
Score = 50.1 bits (118), Expect = 4e-06, Method: Composition-based stats.
Identities = 43/126 (34%), Positives = 65/126 (51%), Gaps = 10/126 (7%)
Query 29 QPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR---------MGGTCV 79
Q +L H +D +V+GGGSGG+A A+ A+ G KVA ++ + +GGTCV
Sbjct 161 QDYLKEWLRDHTYDLIVIGGGSGGLAAAKEASRLGKKVACLDFVKPSPQGTSWGLGGTCV 220
Query 80 NVGCVPKKIMWCAANAFESLQGMKHLGVEFSDPP-RLNWQRLVQNRENYIKRLNRIYEDN 138
NVGC+PKK+M A+ S+ K G + + W L + +++I LN Y
Sbjct 221 NVGCIPKKLMHQASLLGHSIHDAKKYGWKLPEGKVEHQWNHLRDSVQDHIASLNWGYRVQ 280
Query 139 LDKSQV 144
L + V
Sbjct 281 LREKTV 286
> ath:AT1G48030 mtLPD1; mtLPD1 (mitochondrial lipoamide dehydrogenase
1); ATP binding / dihydrolipoyl dehydrogenase; K00382
dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=507
Score = 49.3 bits (116), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 39/143 (27%), Positives = 71/143 (49%), Gaps = 15/143 (10%)
Query 42 DCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAF-ESL 99
D V++GGG GG A +A+ G K +E R +GGTC+NVGC+P K + +++ + E+
Sbjct 45 DVVIIGGGPGGYVAAIKASQLGLKTTCIEKRGALGGTCLNVGCIPSKALLHSSHMYHEAK 104
Query 100 QGMKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLKPKDEA 159
+ G++ S ++ ++ ++N +K L R E K++V G+ +E
Sbjct 105 HSFANHGIKVSS-VEVDLPAMLAQKDNAVKNLTRGIEGLFKKNKVTYVKGYGKFISPNEV 163
Query 160 N------------GQHVVVVNSS 170
+ G+H++V S
Sbjct 164 SVETIDGGNTIVKGKHIIVATGS 186
> eco:b0304 ykgC, ECK0303, JW5040; predicted pyridine nucleotide-disulfide
oxidoreductase
Length=441
Score = 47.4 bits (111), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 2/51 (3%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSR--MGGTCVNVGCVPKKIM 89
+ V++G G G A A G +VAL+E+S GGTC+N+GC+P K +
Sbjct 4 YQAVIIGFGKAGKTLAVTLAKAGWRVALIEQSNAMYGGTCINIGCIPTKTL 54
> tgo:TGME49_006470 dihydrolipoyl dehydrogenase, putative (EC:1.8.1.4);
K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=519
Score = 46.6 bits (109), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 52/100 (52%), Gaps = 3/100 (3%)
Query 56 ARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAF-ESLQGMKHLGVEFSDPP 113
A +AA G K A VE R +GGTC+NVGC+P K + +N + ++ + LG++ D
Sbjct 65 AIKAAQLGLKTACVEKRGTLGGTCLNVGCIPSKAVLNISNKYVDARDHFERLGIKI-DGL 123
Query 114 RLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATL 153
++ ++ + ++ + L + E ++ V Y G L
Sbjct 124 SIDIDKMQKQKQKVVSTLTQGIEHLFRRNGVDYYVGEGKL 163
> sce:YPL017C IRC15; Irc15p; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=499
Score = 44.3 bits (103), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 55/118 (46%), Gaps = 5/118 (4%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVE-RSRMGGTCVNVGCVPKKIMWCAANAFESL 99
+D +V+G G GG A +A+ G A V+ R+ +GG + G VP K + + + L
Sbjct 18 YDVLVIGCGPGGFTAAMQASQAGLLTACVDQRASLGGAYLVDGAVPSKTLLYESYLYRLL 77
Query 100 QG---MKHLGVEFSDPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGFATLK 154
Q ++ G P + + Q ++ I+ L +Y+ L K+ V Y G A K
Sbjct 78 QQQELIEQRGTRLF-PAKFDMQAAQSALKHNIEELGNVYKRELSKNNVTVYKGTAAFK 134
> ath:AT4G16155 dihydrolipoyl dehydrogenase; K00382 dihydrolipoamide
dehydrogenase [EC:1.8.1.4]
Length=630
Score = 43.5 bits (101), Expect = 4e-04, Method: Composition-based stats.
Identities = 31/111 (27%), Positives = 50/111 (45%), Gaps = 5/111 (4%)
Query 59 AATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQG---MKHLGVEFSDPPRL 115
A G K A++E +GGTCVN GCVP K + + LQ MK G++ S
Sbjct 166 AVEKGLKTAIIEGDVVGGTCVNRGCVPSKALLAVSGRMRELQNEHHMKAFGLQVS-AAGY 224
Query 116 NWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF-ATLKPKDEANGQHVV 165
+ Q + + N ++ +++ V GF A L P+ G +++
Sbjct 225 DRQGVADHASNLATKIRNNLTNSMKALGVDILTGFGAVLGPQKVKYGDNII 275
> ath:AT3G16950 LPD1; LPD1 (LIPOAMIDE DEHYDROGENASE 1); dihydrolipoyl
dehydrogenase; K00382 dihydrolipoamide dehydrogenase
[EC:1.8.1.4]
Length=623
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query 59 AATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQG---MKHLGVEFS 110
A G K A++E +GGTCVN GCVP K + + LQ MK G++ S
Sbjct 105 AVEKGLKTAIIEGDVVGGTCVNRGCVPSKALLAVSGRMRELQNEHHMKSFGLQVS 159
> tgo:TGME49_105980 dihydrolipoyl dehydrogenase protein, putative
(EC:1.8.1.4); K00382 dihydrolipoamide dehydrogenase [EC:1.8.1.4]
Length=636
Score = 43.1 bits (100), Expect = 4e-04, Method: Composition-based stats.
Identities = 30/65 (46%), Positives = 36/65 (55%), Gaps = 0/65 (0%)
Query 41 FDCVVLGGGSGGIAFARRAATYGAKVALVERSRMGGTCVNVGCVPKKIMWCAANAFESLQ 100
FD ++G G GG A A AA G K A+V GGTCVN GCVP K + AA + L+
Sbjct 142 FDVTIIGLGVGGHAAALHAAALGLKTAVVSGGDPGGTCVNRGCVPSKALLAAARRVKMLR 201
Query 101 GMKHL 105
HL
Sbjct 202 NKHHL 206
> eco:b3962 sthA, ECK3954, JW5551, sth, udhA; pyridine nucleotide
transhydrogenase, soluble (EC:1.6.1.1); K00322 NAD(P) transhydrogenase
[EC:1.6.1.1]
Length=466
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 34/118 (28%), Positives = 57/118 (48%), Gaps = 16/118 (13%)
Query 35 MAAAHHFDCVVLGGGSGGIAFARRAATYGAKVALVERSR-MGGTCVNVGCVPKKIMWCAA 93
M ++ +D +V+G G GG A GA+VA++ER + +GG C + G +P K + A
Sbjct 1 MPHSYDYDAIVIGSGPGGEGAAMGLVKQGARVAVIERYQNVGGGCTHWGTIPSKALRHAV 60
Query 94 NAFESLQGMKHLGVEFS-DPPRLNWQRLVQNRENYIKRLNRIYEDNLDKSQVHRYYGF 150
+ +EF+ +P + RL+ R ++ LN + DN+ Q GF
Sbjct 61 SRI----------IEFNQNPLYSDHSRLL--RSSFADILN--HADNVINQQTRMRQGF 104
> dre:751628 gpd2, MGC153729, cb853, fe02f02, im:7147983, wu:fe02f02,
zgc:153729; glycerol-3-phosphate dehydrogenase 2 (mitochondrial)
(EC:1.1.5.3); K00111 glycerol-3-phosphate dehydrogenase
[EC:1.1.5.3]
Length=536
Score = 42.4 bits (98), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 23/66 (34%), Positives = 35/66 (53%), Gaps = 2/66 (3%)
Query 9 KCCFLSLQQQLQQ--RDSAPASQPHLSAMAAAHHFDCVVLGGGSGGIAFARRAATYGAKV 66
+ ++ Q +L+ +D+ P + LS + FD +V+GGG+ G A A T K
Sbjct 35 RLAYVEAQGELKVPFKDALPTREEQLSTLRNTEEFDVLVVGGGATGSGCALDAVTRNLKT 94
Query 67 ALVERS 72
ALVERS
Sbjct 95 ALVERS 100
Lambda K H
0.321 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4535951560
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40