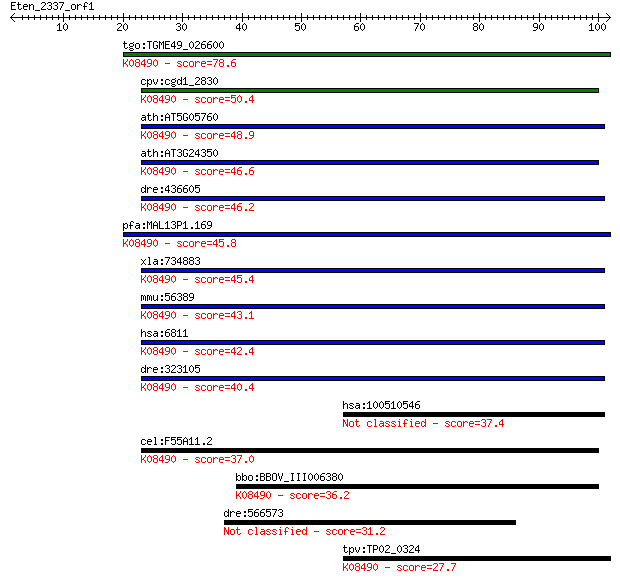
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2337_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5 78.6
cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane dom... 50.4 1e-06
ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP recep... 48.9 4e-06
ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP recep... 46.6 2e-05
dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490 s... 46.2 2e-05
pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative; K0... 45.8 3e-05
xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5 45.4
mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin 5A... 43.1 2e-04
hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5 42.4
dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like; K... 40.4 0.002
hsa:100510546 syntaxin-5-like 37.4 0.012
cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syn... 37.0 0.017
bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syn... 36.2 0.026
dre:566573 fam65a, si:dkey-45h7.2; family with sequence simila... 31.2 0.85
tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5 27.7
> tgo:TGME49_026600 syntaxin, putative ; K08490 syntaxin 5
Length=283
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 44/83 (53%), Positives = 56/83 (67%), Gaps = 1/83 (1%)
Query 20 MAYDRTAEFAAAAESFNFAASPPAADLR-QPFSDADKNFNLIASDMGNSLHATSLKLQEL 78
M DRTA+F A AE + A A +LR + AD +FN A+++G LH TSLKL+EL
Sbjct 1 MPCDRTADFLAFAERASPGAISQARELRSRTVRHADNSFNASAAEIGTQLHRTSLKLKEL 60
Query 79 AKLARQCGIYNDKTAQIQELTFE 101
AK ARQ IYND+TAQ Q+LT+E
Sbjct 61 AKFARQRSIYNDRTAQTQDLTYE 83
> cpv:cgd1_2830 syntaxin 5A ortholog, possible transmembrane domain
or GPI at C-terminus ; K08490 syntaxin 5
Length=329
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 27/88 (30%), Positives = 47/88 (53%), Gaps = 17/88 (19%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADK-----------NFNLIASDMGNSLHAT 71
DRTA+F FNF + ++ + FNL+AS++ +++T
Sbjct 10 DRTADF------FNFVEKHDSTRSEGRIGNSGQSNRYSNSIQGSQFNLLASEISQEMNST 63
Query 72 SLKLQELAKLARQCGIYNDKTAQIQELT 99
SLK++EL ++ +Q G++ D+T QI +LT
Sbjct 64 SLKIEELNRIVKQKGLFRDRTNQIHQLT 91
> ath:AT5G05760 SYP31; SYP31 (SYNTAXIN OF PLANTS 31); SNAP receptor;
K08490 syntaxin 5
Length=336
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 33/87 (37%), Positives = 45/87 (51%), Gaps = 12/87 (13%)
Query 23 DRTAEFAAAAESFN-FAASP--------PAADLRQPFSDADKNFNLIASDMGNSLHATSL 73
DRT E + +++ A P PA+ R S FN AS +G + TS
Sbjct 7 DRTVELHSLSQTLKKIGAIPSVHQDEDDPASSKR---SSPGSEFNKKASRIGLGIKETSQ 63
Query 74 KLQELAKLARQCGIYNDKTAQIQELTF 100
K+ LAKLA+Q I+ND+T +IQELT
Sbjct 64 KITRLAKLAKQSTIFNDRTVEIQELTV 90
> ath:AT3G24350 SYP32; SYP32 (SYNTAXIN OF PLANTS 32); SNAP receptor;
K08490 syntaxin 5
Length=347
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 14/90 (15%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADKN-------------FNLIASDMGNSLH 69
DR+ EF E+ + +P A P+ + ++N FN AS +G +++
Sbjct 12 DRSDEFFKIVETLRRSIAPAPAANNVPYGN-NRNDGARREDLINKSEFNKRASHIGLAIN 70
Query 70 ATSLKLQELAKLARQCGIYNDKTAQIQELT 99
TS KL +LAKLA++ +++D T +IQELT
Sbjct 71 QTSQKLSKLAKLAKRTSVFDDPTQEIQELT 100
> dre:436605 stx5a, wu:fj81f08, zgc:91864; syntaxin 5A; K08490
syntaxin 5
Length=302
Score = 46.2 bits (108), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 46/84 (54%), Gaps = 10/84 (11%)
Query 23 DRTAEFAAAAESF------NFAASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQ 76
DRT EF +A +S N S PA + + SD F L+A +G L T KL+
Sbjct 5 DRTLEFQSACKSLQGRQLQNGTHSKPANNALKQRSD----FTLMAKRIGKDLSNTFAKLE 60
Query 77 ELAKLARQCGIYNDKTAQIQELTF 100
+L LA++ +++DK +I+ELT+
Sbjct 61 KLTILAKRKSLFDDKAVEIEELTY 84
> pfa:MAL13P1.169 PfSyn5, PfStx5; Qa-SNARE protein, putative;
K08490 syntaxin 5
Length=281
Score = 45.8 bits (107), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 30/84 (35%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query 20 MAY-DRTAEFAAAAESFNFAASPPAADLRQPFSDA-DKNFNLIASDMGNSLHATSLKLQE 77
M Y D+T EF E S ++R+ S D +AS + + L + KLQ+
Sbjct 1 MPYVDKTEEFFKIIEKL----SNDNINIRKNRSIVQDTQVGELASKITDLLQSGYQKLQQ 56
Query 78 LAKLARQCGIYNDKTAQIQELTFE 101
L + +Q GI+NDKT++I+ELT+E
Sbjct 57 LERCVKQKGIFNDKTSEIEELTYE 80
> xla:734883 stx5, MGC114979; syntaxin 5; K08490 syntaxin 5
Length=298
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/81 (35%), Positives = 44/81 (54%), Gaps = 5/81 (6%)
Query 23 DRTAEFAAAAESFNFAASPPAADLRQPFSDADKN---FNLIASDMGNSLHATSLKLQELA 79
DRTAEF + +S L P +A K F L+A +G L T KL++L
Sbjct 5 DRTAEFISTCKSLQ--GRQNGVQLSSPSLNAVKQRSEFTLMAKRIGKDLSNTFSKLEKLT 62
Query 80 KLARQCGIYNDKTAQIQELTF 100
LA++ +++DK A+I+ELT+
Sbjct 63 ILAKRKSLFDDKAAEIEELTY 83
> mmu:56389 Stx5a, 0610031F24Rik, D19Ertd627e, Stx5; syntaxin
5A; K08490 syntaxin 5
Length=355
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/83 (33%), Positives = 44/83 (53%), Gaps = 9/83 (10%)
Query 23 DRTAEFAAAAESF-----NFAASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQE 77
DRT EF +A +S S PA + S+ F L+A +G L T KL++
Sbjct 59 DRTQEFQSACKSLQSRQNGIQTSKPALHAARQCSE----FTLMARRIGKDLSNTFAKLEK 114
Query 78 LAKLARQCGIYNDKTAQIQELTF 100
L LA++ +++DK +I+ELT+
Sbjct 115 LTILAKRKSLFDDKAVEIEELTY 137
> hsa:6811 STX5, SED5, STX5A; syntaxin 5; K08490 syntaxin 5
Length=355
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 44/83 (53%), Gaps = 9/83 (10%)
Query 23 DRTAEFAAAAESF-----NFAASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQE 77
DRT EF +A +S + PA + S+ F L+A +G L T KL++
Sbjct 59 DRTQEFLSACKSLQTRQNGIQTNKPALRAVRQRSE----FTLMAKRIGKDLSNTFAKLEK 114
Query 78 LAKLARQCGIYNDKTAQIQELTF 100
L LA++ +++DK +I+ELT+
Sbjct 115 LTILAKRKSLFDDKAVEIEELTY 137
> dre:323105 stx5al, wu:fb81g12, zgc:73276; syntaxin 5A, like;
K08490 syntaxin 5
Length=298
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 43/83 (51%), Gaps = 9/83 (10%)
Query 23 DRTAEFAAAAESFNF---AASPPAA--DLRQPFSDADKNFNLIASDMGNSLHATSLKLQE 77
DRT EF + +S A P A + Q SD F L+A +G L T KL++
Sbjct 5 DRTGEFQSVCKSLQGRQNGAQPVRAVNNAIQKRSD----FTLLAKRIGRDLSNTFAKLEK 60
Query 78 LAKLARQCGIYNDKTAQIQELTF 100
L LA++ +++DK +I ELT+
Sbjct 61 LTILAKRKSLFDDKATEIDELTY 83
> hsa:100510546 syntaxin-5-like
Length=295
Score = 37.4 bits (85), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 29/44 (65%), Gaps = 0/44 (0%)
Query 57 FNLIASDMGNSLHATSLKLQELAKLARQCGIYNDKTAQIQELTF 100
F L+A +G L T KL++L LA++ +++DK +I+ELT+
Sbjct 34 FTLMAKRIGKDLSNTFAKLEKLTILAKRKSLFDDKAVEIEELTY 77
> cel:F55A11.2 syn-3; SYNtaxin family member (syn-3); K08490 syntaxin
5
Length=413
Score = 37.0 bits (84), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 27/81 (33%), Positives = 45/81 (55%), Gaps = 7/81 (8%)
Query 23 DRTAEFAAAAESFNFAASPPAADLR-QPFSDA---DKNFNLIASDMGNSLHATSLKLQEL 78
DRT+EF A A+S+ A+ A +R QP + FN +A +G L T K+++L
Sbjct 111 DRTSEFRATAKSYEMKAA--ANGIRPQPKHEMLSESVQFNQLAKRIGKELSQTCAKMEKL 168
Query 79 AKLARQCGIYNDKTAQIQELT 99
A+ A++ Y +++ QI L+
Sbjct 169 AEYAKKKSCYEERS-QIDHLS 188
> bbo:BBOV_III006380 17.m07568; hypothetical protein; K08490 syntaxin
5
Length=256
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/61 (39%), Positives = 33/61 (54%), Gaps = 8/61 (13%)
Query 39 ASPPAADLRQPFSDADKNFNLIASDMGNSLHATSLKLQELAKLARQCGIYNDKTAQIQEL 98
A+PPAA+ Q +A + +G L KL EL+ LAR+ IY D TA+I+ L
Sbjct 26 AAPPAAESSQLEQEAQR--------VGLQLSKCETKLTELSALARKRSIYVDHTAEIERL 77
Query 99 T 99
T
Sbjct 78 T 78
> dre:566573 fam65a, si:dkey-45h7.2; family with sequence similarity
65, member A
Length=1082
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 31/56 (55%), Gaps = 7/56 (12%)
Query 37 FAASPPAADLRQPFSDADKNFN-------LIASDMGNSLHATSLKLQELAKLARQC 85
+ ASP + + R+ S+A+K+F ++ S++ N L +K++ LA AR C
Sbjct 161 YTASPGSREARESLSEANKSFKEYTENMCMLESELENQLGEFHIKMKGLAGFARLC 216
> tpv:TP02_0324 syntaxin 5; K08490 syntaxin 5
Length=285
Score = 27.7 bits (60), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 0/45 (0%)
Query 57 FNLIASDMGNSLHATSLKLQELAKLARQCGIYNDKTAQIQELTFE 101
FN A + + KL EL++LA++ +Y D T+ I+ LT E
Sbjct 35 FNTDADSIYKEIEKARAKLNELSQLAKKRSLYLDNTSSIERLTSE 79
Lambda K H
0.307 0.116 0.303
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2036602604
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40