bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2288_orf2
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
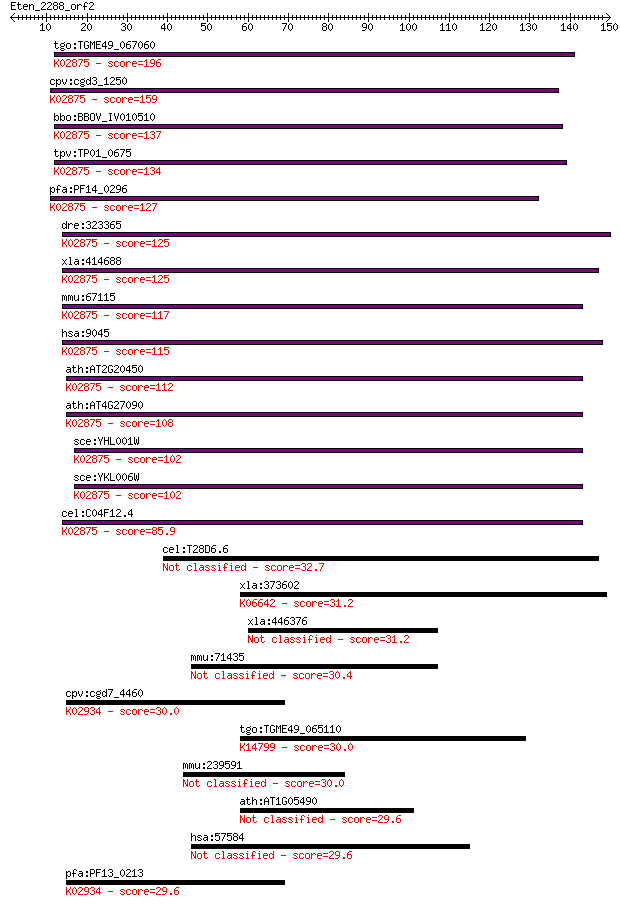
tgo:TGME49_067060 60S ribosomal protein L14, putative ; K02875... 196 2e-50
cpv:cgd3_1250 ribosomal protein L14 ; K02875 large subunit rib... 159 3e-39
bbo:BBOV_IV010510 23.m06087; ribosomal protein L14; K02875 lar... 137 1e-32
tpv:TP01_0675 60S ribosomal protein L14; K02875 large subunit ... 134 9e-32
pfa:PF14_0296 60S ribosomal protein L14, putative; K02875 larg... 127 1e-29
dre:323365 rpl14, MGC111817, wu:fb96c03, zgc:111817; ribosomal... 125 6e-29
xla:414688 rpl14, MGC83076; ribosomal protein L14; K02875 larg... 125 6e-29
mmu:67115 Rpl14, 3100001N19Rik, AA407502, AL022816, CAG-ISL-7,... 117 2e-26
hsa:9045 RPL14, CAG-ISL-7, CTG-B33, L14, MGC88594, RL14, hRL14... 115 4e-26
ath:AT2G20450 60S ribosomal protein L14 (RPL14A); K02875 large... 112 3e-25
ath:AT4G27090 60S ribosomal protein L14 (RPL14B); K02875 large... 108 4e-24
sce:YHL001W RPL14B; L14B; K02875 large subunit ribosomal prote... 102 6e-22
sce:YKL006W RPL14A; L14A; K02875 large subunit ribosomal prote... 102 6e-22
cel:C04F12.4 rpl-14; Ribosomal Protein, Large subunit family m... 85.9 4e-17
cel:T28D6.6 hypothetical protein 32.7 0.49
xla:373602 prkdc, dna-pkcs, dnapk, dnpk1, hyrc, hyrc1, p350, p... 31.2 1.2
xla:446376 arhgap21, MGC83679, arhgap21-A; Rho GTPase activati... 31.2 1.2
mmu:71435 Arhgap21, 5530401C11Rik, AA416458, ARHGAP10; Rho GTP... 30.4 2.1
cpv:cgd7_4460 60S ribosomal protein L6 ; K02934 large subunit ... 30.0 2.6
tgo:TGME49_065110 hypothetical protein ; K14799 pre-rRNA-proce... 30.0 2.7
mmu:239591 Ttll8, 1700019P01Rik, 4933401B17; tubulin tyrosine ... 30.0 3.0
ath:AT1G05490 chr31; chr31 (chromatin remodeling 31); ATP bind... 29.6 3.3
hsa:57584 ARHGAP21, ARHGAP10, DKFZp761L0424, FLJ33323, FLJ9010... 29.6 3.6
pfa:PF13_0213 60S ribosomal protein L6-2, putative; K02934 lar... 29.6 4.1
> tgo:TGME49_067060 60S ribosomal protein L14, putative ; K02875
large subunit ribosomal protein L14e
Length=134
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 90/129 (69%), Positives = 110/129 (85%), Gaps = 0/129 (0%)
Query 12 MALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALT 71
M LF R+ +PGRLCVVQYGPD GKLCFVVDIIN R+LVDGA VTGV+RQS+P+RR+ALT
Sbjct 1 MGLFTRYYQPGRLCVVQYGPDAGKLCFVVDIINQTRVLVDGAGVTGVKRQSMPVRRIALT 60
Query 72 DMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQ 131
D LKIP VRS TL+KALEKDDVI KFN+SSWG++R+AK++RA ++DF+RFKLMV KQ
Sbjct 61 DQYLKIPRSVRSATLKKALEKDDVIAKFNQSSWGKRRLAKEQRANMSDFDRFKLMVILKQ 120
Query 132 RAQAVRAKL 140
R + ++ KL
Sbjct 121 RRKVMQQKL 129
> cpv:cgd3_1250 ribosomal protein L14 ; K02875 large subunit ribosomal
protein L14e
Length=125
Score = 159 bits (403), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 73/126 (57%), Positives = 99/126 (78%), Gaps = 1/126 (0%)
Query 11 KMALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVAL 70
KM LF RF+EPGRLC +QYGPD GK+CF++D+IN NRIL+DG + T V RQSIP++R+AL
Sbjct 1 KMTLFTRFVEPGRLCRIQYGPDTGKMCFIIDVINMNRILIDGPS-TNVARQSIPLKRLAL 59
Query 71 TDMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARK 130
TD + KIP G R+ T++K LEKD+ I+ FN++++G+K AK +A +TDFER L+VARK
Sbjct 60 TDFKAKIPRGARTGTVKKILEKDNTIENFNKTTYGQKCAAKIFKANMTDFERHALLVARK 119
Query 131 QRAQAV 136
+R V
Sbjct 120 KRQYLV 125
> bbo:BBOV_IV010510 23.m06087; ribosomal protein L14; K02875 large
subunit ribosomal protein L14e
Length=133
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 62/126 (49%), Positives = 89/126 (70%), Gaps = 0/126 (0%)
Query 12 MALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALT 71
M LF++F+EPGRLC++ YGPD GKLC +VDI+ RILVDG +TGV RQ IP+ + LT
Sbjct 1 MPLFKKFVEPGRLCLITYGPDAGKLCVIVDIVTNTRILVDGGMITGVERQQIPVTWIKLT 60
Query 72 DMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQ 131
D ++ + G ++ T+ KA++K D + F ++ G+K +RR LTDFERFKL VA+K+
Sbjct 61 DTKITLNRGAKTGTVGKAVKKFDALNAFKQTPLGKKLEIAERRKNLTDFERFKLSVAQKE 120
Query 132 RAQAVR 137
R + +R
Sbjct 121 RRRLMR 126
> tpv:TP01_0675 60S ribosomal protein L14; K02875 large subunit
ribosomal protein L14e
Length=130
Score = 134 bits (338), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 60/127 (47%), Positives = 94/127 (74%), Gaps = 0/127 (0%)
Query 12 MALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALT 71
M LF++F+EPGRLC++ YGP+ GKL FVVDI+ R+LVDGA+VTGV+RQ +P + LT
Sbjct 1 MPLFKKFVEPGRLCLLTYGPNSGKLVFVVDIVTPTRVLVDGADVTGVKRQQVPKTWLKLT 60
Query 72 DMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQ 131
D+R+++ G R+ TL KA + + ++ F+++S G+K +++ L DF+RFKLM A+KQ
Sbjct 61 DVRVELQRGARTGTLSKAAKAQNALELFDKTSLGKKLRVLEKKKNLNDFQRFKLMSAKKQ 120
Query 132 RAQAVRA 138
R + +++
Sbjct 121 RRKVLKS 127
> pfa:PF14_0296 60S ribosomal protein L14, putative; K02875 large
subunit ribosomal protein L14e
Length=165
Score = 127 bits (320), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 58/121 (47%), Positives = 85/121 (70%), Gaps = 0/121 (0%)
Query 11 KMALFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVAL 70
K LF+RF+EPGRLC+++YGP GKLCF+VDI+ R++VDGA +TGV R IP++R+ L
Sbjct 17 KNLLFKRFVEPGRLCLIEYGPYAGKLCFIVDIVTITRVIVDGAFITGVPRMVIPLKRLKL 76
Query 71 TDMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARK 130
R+KI +S LRK + V+++FN S G+K I K++R TDFERF++ A++
Sbjct 77 LKERIKINKNCKSGFLRKTVNSTKVLEEFNNSKLGKKMIIKKKRDEATDFERFQVYFAKR 136
Query 131 Q 131
+
Sbjct 137 E 137
> dre:323365 rpl14, MGC111817, wu:fb96c03, zgc:111817; ribosomal
protein L14; K02875 large subunit ribosomal protein L14e
Length=139
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 60/136 (44%), Positives = 98/136 (72%), Gaps = 2/136 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+F+RF+E GR+ + +GP EGKL +VD+I+ NR LVDG TGV+RQ++P + + LTD
Sbjct 2 VFKRFVEIGRVAFIAFGPHEGKLVAIVDVIDQNRALVDGP-CTGVKRQALPFKCLQLTDY 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
+K+PH R +R+A EK ++ KK+ ESSW +K A+++RAA++DF+R+K+ A+K R
Sbjct 61 VIKVPHRNRQKYVRRAWEKAEISKKWEESSWAKKIEARKKRAAMSDFDRYKVSKAKKMRN 120
Query 134 QAVRAKLGKRMQKVES 149
+ +R ++ K++QK +
Sbjct 121 KIIRHEM-KKLQKAAA 135
> xla:414688 rpl14, MGC83076; ribosomal protein L14; K02875 large
subunit ribosomal protein L14e
Length=138
Score = 125 bits (313), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 60/133 (45%), Positives = 96/133 (72%), Gaps = 2/133 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+F+R+++ GR+ + +GP GKL +VD+I+ NR LVDG TGVRRQ++P + + LTD
Sbjct 2 VFKRYVQIGRVAYLSFGPHAGKLVAIVDVIDQNRALVDGP-CTGVRRQAMPFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
+K+PH R +R+A EK +V +K+ ESSW +K A+QR+A ++DF+RFK+M A+K R
Sbjct 61 VIKVPHSARQKYVRRAWEKANVNEKWAESSWAKKIDARQRKAKMSDFDRFKVMKAKKMRN 120
Query 134 QAVRAKLGKRMQK 146
+ +R ++ K++QK
Sbjct 121 KIIRHEM-KKLQK 132
> mmu:67115 Rpl14, 3100001N19Rik, AA407502, AL022816, CAG-ISL-7,
CTG-B33, L14; ribosomal protein L14; K02875 large subunit
ribosomal protein L14e
Length=217
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 57/129 (44%), Positives = 87/129 (67%), Gaps = 1/129 (0%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+FRR++E GR+ + +GP GKL +VD+I+ NR LVDG T VRRQ++P + + LTD
Sbjct 2 VFRRYVEVGRVAYISFGPHAGKLVAIVDVIDQNRALVDGP-CTRVRRQAMPFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK PH R +RKA EK D+ K+ + W +K A++R+A +TDF+RFK+M A+K R
Sbjct 61 ILKFPHSARQKYVRKAWEKADINTKWAATRWAKKIDARERKAKMTDFDRFKVMKAKKMRN 120
Query 134 QAVRAKLGK 142
+ ++ ++ K
Sbjct 121 RIIKTEVKK 129
> hsa:9045 RPL14, CAG-ISL-7, CTG-B33, L14, MGC88594, RL14, hRL14;
ribosomal protein L14; K02875 large subunit ribosomal protein
L14e
Length=215
Score = 115 bits (289), Expect = 4e-26, Method: Compositional matrix adjust.
Identities = 58/134 (43%), Positives = 90/134 (67%), Gaps = 2/134 (1%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+FRRF+E GR+ V +GP GKL +VD+I+ NR LVDG T VRRQ++P + + LTD
Sbjct 2 VFRRFVEVGRVAYVSFGPHAGKLVAIVDVIDQNRALVDGP-CTQVRRQAMPFKCMQLTDF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK PH +R+A +K D+ K+ + W +K A++R+A +TDF+RFK+M A+K R
Sbjct 61 ILKFPHSAHQKYVRQAWQKADINTKWAATRWAKKIEARERKAKMTDFDRFKVMKAKKMRN 120
Query 134 QAVRAKLGKRMQKV 147
+ ++ ++ K++QK
Sbjct 121 RIIKNEV-KKLQKA 133
> ath:AT2G20450 60S ribosomal protein L14 (RPL14A); K02875 large
subunit ribosomal protein L14e
Length=134
Score = 112 bits (281), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 58/128 (45%), Positives = 88/128 (68%), Gaps = 3/128 (2%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMR 74
F+RF+E GR+ +V YG D GKL +VD+++ NR LVD ++ R + ++R++LTD+
Sbjct 3 FKRFVEIGRVALVNYGEDYGKLVVIVDVVDQNRALVDAPDME---RIQMNLKRLSLTDIV 59
Query 75 LKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQ 134
+ I + L +A+EK DV K+ +SSWGRK I ++RRAAL DF+RFK+M+A+ +RA
Sbjct 60 IDINRVPKKKVLIEAMEKADVKNKWEKSSWGRKLIVQKRRAALNDFDRFKIMLAKIKRAG 119
Query 135 AVRAKLGK 142
VR +L K
Sbjct 120 IVRQELAK 127
> ath:AT4G27090 60S ribosomal protein L14 (RPL14B); K02875 large
subunit ribosomal protein L14e
Length=134
Score = 108 bits (271), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 55/128 (42%), Positives = 86/128 (67%), Gaps = 3/128 (2%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMR 74
F+R++E GR+ +V YG D GKL +VD+++ NR LVD ++ R + +R++LTD+
Sbjct 3 FKRYVEIGRVALVNYGEDHGKLVVIVDVVDQNRALVDAPDM---ERIQMNFKRLSLTDIV 59
Query 75 LKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQ 134
+ I + L +A+EK DV K+ +SSWGRK I ++RRA L DF+RFK+M+A+ ++A
Sbjct 60 IDINRVPKKKALIEAMEKADVKNKWEKSSWGRKLIVQKRRANLNDFDRFKIMLAKIKKAG 119
Query 135 AVRAKLGK 142
VR +L K
Sbjct 120 VVRQELAK 127
> sce:YHL001W RPL14B; L14B; K02875 large subunit ribosomal protein
L14e
Length=138
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 52/126 (41%), Positives = 79/126 (62%), Gaps = 1/126 (0%)
Query 17 RFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLK 76
R +E GR+ +++ G GKL +V+II+ ++L+DG GV RQ+I + +V LT +
Sbjct 13 RLVEVGRVVLIKKGQSAGKLAAIVEIIDQKKVLIDGPK-AGVPRQAINLGQVVLTPLTFA 71
Query 77 IPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAV 136
+P G R+ T+ K V +K+ SSW +K ++RRAALTDFERF++MV RKQ+ V
Sbjct 72 LPRGARTATVSKKWAAAGVCEKWAASSWAKKIAQRERRAALTDFERFQVMVLRKQKRYTV 131
Query 137 RAKLGK 142
+ L K
Sbjct 132 KKALAK 137
> sce:YKL006W RPL14A; L14A; K02875 large subunit ribosomal protein
L14e
Length=138
Score = 102 bits (253), Expect = 6e-22, Method: Compositional matrix adjust.
Identities = 52/126 (41%), Positives = 79/126 (62%), Gaps = 1/126 (0%)
Query 17 RFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLK 76
R +E GR+ +++ G GKL +V+II+ ++L+DG GV RQ+I + +V LT +
Sbjct 13 RLVEVGRVVLIKKGQSAGKLAAIVEIIDQKKVLIDGPK-AGVPRQAINLGQVVLTPLTFA 71
Query 77 IPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAV 136
+P G R+ T+ K V +K+ SSW +K ++RRAALTDFERF++MV RKQ+ V
Sbjct 72 LPRGARTATVSKKWAAAAVCEKWAASSWAKKIAQRERRAALTDFERFQVMVLRKQKRYTV 131
Query 137 RAKLGK 142
+ L K
Sbjct 132 KKALAK 137
> cel:C04F12.4 rpl-14; Ribosomal Protein, Large subunit family
member (rpl-14); K02875 large subunit ribosomal protein L14e
Length=135
Score = 85.9 bits (211), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 46/129 (35%), Positives = 78/129 (60%), Gaps = 1/129 (0%)
Query 14 LFRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRVALTDM 73
+F R ++ GR+ + G D+GKL +V++I+GNR+ +DG + + V R ++ + LT
Sbjct 2 VFNRVVQIGRVVFIASGKDQGKLAAIVNVIDGNRVQIDGPS-SDVTRTVRNLKDLQLTKF 60
Query 74 RLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAALTDFERFKLMVARKQRA 133
LK+ G R+ ++ A + V + F ++ W +K + RA LTDFER+KLM A++ R
Sbjct 61 VLKLRVGQRTKGVKAAFDAAKVTENFQKTQWAKKIAQRAIRAKLTDFERYKLMKAKQMRN 120
Query 134 QAVRAKLGK 142
+ VR +L K
Sbjct 121 RIVRVELAK 129
> cel:T28D6.6 hypothetical protein
Length=366
Score = 32.7 bits (73), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 27/108 (25%), Positives = 52/108 (48%), Gaps = 6/108 (5%)
Query 39 VVDIINGNRILVDGANVTGVRRQSIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKK 98
++D+I GNRI + V + I I + D+ +IPH V ++ DD+++K
Sbjct 228 LIDVIEGNRIYIPCIYVLN-KIDQISIEEL---DIIYRIPHTV-PISAHHKWNFDDLLEK 282
Query 99 FNESSWGRKRIAKQRRAALTDFERFKLMVARKQRAQAVRAKLGKRMQK 146
E RI + + L D+ + ++ A ++ + + K+ K +QK
Sbjct 283 VWE-YLNLIRIYTKPKGQLPDYSQPIVLNAERKSIEDLCTKIHKSLQK 329
> xla:373602 prkdc, dna-pkcs, dnapk, dnpk1, hyrc, hyrc1, p350,
prkdc-A, xrcc7; protein kinase, DNA-activated, catalytic polypeptide
(EC:2.7.11.1); K06642 DNA-dependent protein kinase
catalytic subunit [EC:2.7.11.1]
Length=4146
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 42/91 (46%), Gaps = 7/91 (7%)
Query 58 VRRQSIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGRKRIAKQRRAAL 117
V +++ +RRV L + G+R + L DV +S R I + RR L
Sbjct 2706 VNKRNENVRRVPLKPLGPNF--GMRRLGL-----PGDVTDSKTKSMEDRSDILRLRRRFL 2758
Query 118 TDFERFKLMVARKQRAQAVRAKLGKRMQKVE 148
D E+ L+ ARK A+ R K K QK++
Sbjct 2759 KDREKLSLIYARKGTAEQKREKAIKIEQKMK 2789
> xla:446376 arhgap21, MGC83679, arhgap21-A; Rho GTPase activating
protein 21
Length=1926
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 60 RQSIPIR---RVALTDMRLKIPHGVRSVTLRKALEKDDVIKKFNESSWGR 106
RQ++ IR R D +L+ PH + + R+ KDD+ ++ SW R
Sbjct 1059 RQTLSIRQPFRATRPDGKLQSPHSPKQESERRLFSKDDISPPKDKGSWRR 1108
> mmu:71435 Arhgap21, 5530401C11Rik, AA416458, ARHGAP10; Rho GTPase
activating protein 21
Length=1945
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query 46 NRILVDGANVTGVRRQSIPIRRVAL---TDMRLKIPHGVRSVTLRKALEKDDVIKKFNES 102
N +L + RQS+ IR+ L ++ + + PH + + RK L KDD ++
Sbjct 1056 NSLLSKTEQLPKTPRQSLSIRQTLLGAKSEPKTQSPHSPKEESERKLLSKDDTSPPKDKG 1115
Query 103 SWGR 106
+W R
Sbjct 1116 TWRR 1119
> cpv:cgd7_4460 60S ribosomal protein L6 ; K02934 large subunit
ribosomal protein L6e
Length=184
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 14/54 (25%), Positives = 27/54 (50%), Gaps = 4/54 (7%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGANVTGVRRQSIPIRRV 68
R ++PG +C++ GP +GK ++ + ++V G +P+RRV
Sbjct 35 LRSSLQPGVVCILLAGPYKGKRVVMLKALPSGLVVVTGPYALN----GVPVRRV 84
> tgo:TGME49_065110 hypothetical protein ; K14799 pre-rRNA-processing
protein TSR1
Length=1007
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Query 58 VRRQSIPIRRVALTDMRLKIPHGVRS---VTLRKALEKDDVIKKFNESSWGRKRIAKQRR 114
V++++ I + A D L+ P V + V R +K +K F S W +
Sbjct 665 VKKRNFTIEQRAAED--LQFPDEVDTPLDVEARVRFQKYRGLKSFRTSYWNKDEDLPVDY 722
Query 115 AALTDFERFKLMVA 128
A + DFERFK M A
Sbjct 723 ARVVDFERFKAMSA 736
> mmu:239591 Ttll8, 1700019P01Rik, 4933401B17; tubulin tyrosine
ligase-like family, member 8
Length=832
Score = 30.0 bits (66), Expect = 3.0, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 44 NGNRILVDGANVTGVRRQSIPIRRVALTDMRLKIPHGVRS 83
NG+ + V+G +V ++Q PI V L++ L P VRS
Sbjct 641 NGSDLCVEGISVKKAKKQMPPIASVGLSESLLDAPPKVRS 680
> ath:AT1G05490 chr31; chr31 (chromatin remodeling 31); ATP binding
/ DNA binding / helicase/ nucleic acid binding
Length=1410
Score = 29.6 bits (65), Expect = 3.3, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 0/43 (0%)
Query 58 VRRQSIPIRRVALTDMRLKIPHGVRSVTLRKALEKDDVIKKFN 100
+R+ S PI+RV+L + + + + +L K E+D+ I+K N
Sbjct 321 LRKASSPIKRVSLVERKALVRYKRSGSSLTKPRERDNKIQKLN 363
> hsa:57584 ARHGAP21, ARHGAP10, DKFZp761L0424, FLJ33323, FLJ90108;
Rho GTPase activating protein 21
Length=1958
Score = 29.6 bits (65), Expect = 3.6, Method: Composition-based stats.
Identities = 20/72 (27%), Positives = 34/72 (47%), Gaps = 4/72 (5%)
Query 46 NRILVDGANVTGVRRQSIPIRRVAL---TDMRLKIPHGVRSVTLRKALEKDDVIKKFNES 102
N ++ + RQS+ IR+ L ++ + + PH + + RK L KDD ++
Sbjct 1062 NNLMSKAEQLPKTPRQSLSIRQTLLGAKSEPKTQSPHSPKEESERKLLSKDDTSPPKDKG 1121
Query 103 SWGRKRIAKQRR 114
+W RK I R
Sbjct 1122 TW-RKGIPSIMR 1132
> pfa:PF13_0213 60S ribosomal protein L6-2, putative; K02934 large
subunit ribosomal protein L6e
Length=221
Score = 29.6 bits (65), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 6/55 (10%)
Query 15 FRRFIEPGRLCVVQYGPDEGKLCFVVDIINGNRILVDGA-NVTGVRRQSIPIRRV 68
R+ IE G++ ++ G GK C + I+N + V G + GV P++RV
Sbjct 50 LRKSIEVGKVAIILTGKHMGKRCIITKILNSGLLAVVGPYEINGV-----PLKRV 99
Lambda K H
0.325 0.139 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3068761412
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40