bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
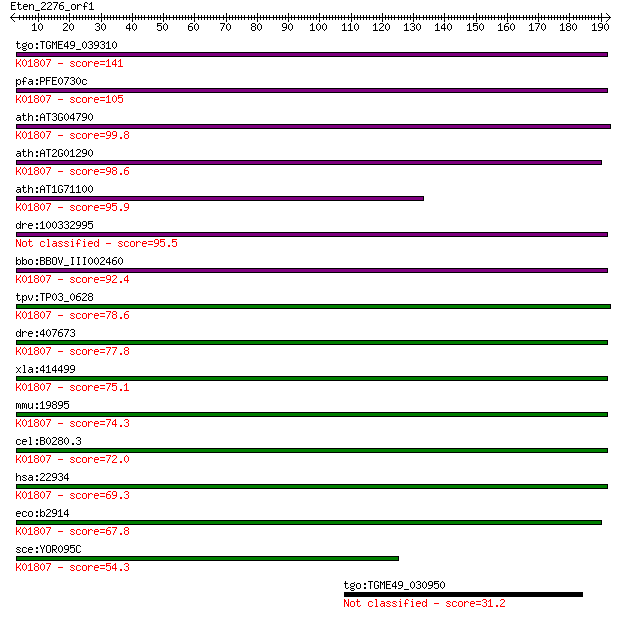
Query= Eten_2276_orf1
Length=192
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_039310 ribose 5-phosphate isomerase, putative (EC:5... 141 2e-33
pfa:PFE0730c ribose 5-phosphate epimerase, putative (EC:5.3.1.... 105 9e-23
ath:AT3G04790 ribose 5-phosphate isomerase-related; K01807 rib... 99.8 6e-21
ath:AT2G01290 ribose-5-phosphate isomerase (EC:5.3.1.6); K0180... 98.6 1e-20
ath:AT1G71100 RSW10; RSW10 (RADIAL SWELLING 10); ribose-5-phos... 95.9 8e-20
dre:100332995 chloroplast ribose-5-phosphate isomerase-like 95.5 9e-20
bbo:BBOV_III002460 17.m07239; ribose 5-phosphate isomerase A f... 92.4 8e-19
tpv:TP03_0628 ribose 5-phosphate epimerase (EC:5.3.1.6); K0180... 78.6 1e-14
dre:407673 rpia, MGC103524, zgc:103524; ribose 5-phosphate iso... 77.8 2e-14
xla:414499 rpia, MGC83218; ribose 5-phosphate isomerase A (EC:... 75.1 1e-13
mmu:19895 Rpia, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.... 74.3 2e-13
cel:B0280.3 hypothetical protein; K01807 ribose 5-phosphate is... 72.0 1e-12
hsa:22934 RPIA, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.... 69.3 8e-12
eco:b2914 rpiA, ECK2910, JW5475, ygfC; ribose 5-phosphate isom... 67.8 2e-11
sce:YOR095C RKI1; Ribose-5-phosphate ketol-isomerase, catalyze... 54.3 2e-07
tgo:TGME49_030950 hypothetical protein 31.2 2.1
> tgo:TGME49_039310 ribose 5-phosphate isomerase, putative (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=259
Score = 141 bits (355), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 90/191 (47%), Positives = 121/191 (63%), Gaps = 11/191 (5%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTG+TA F VER+ +R+++G L D+ C TSEATRK AESLGI + +LD I+ LDV I
Sbjct 28 LGTGTTAKFVVERIGQRMQEGSLKDLLCVPTSEATRKQAESLGIPLTTLDGIADCLDVAI 87
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADE+L + L+KGRGGALLREK++ +K FI ADE K+V + G+TGA+ +E
Sbjct 88 DGADEILP--PTLGLVKGRGGALLREKMIAAAAKTFIVAADETKLVS-NGIGSTGALPVE 144
Query 123 VVQFGAQATRQAVLSAV--VSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVS 180
VV F T++ +LSA+ V GG E + E+ G R E+R FV+
Sbjct 145 VVVFSGSHTKR-LLSALPSVKRHGGRA-EFRKRAGAATGEKNGGQEDIR-EEDR---FVT 198
Query 181 DNGNLCLDLFF 191
DNGN +DL+F
Sbjct 199 DNGNYIVDLYF 209
> pfa:PFE0730c ribose 5-phosphate epimerase, putative (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=236
Score = 105 bits (262), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 67/189 (35%), Positives = 102/189 (53%), Gaps = 27/189 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGST + +ER+ ++ G+L D+ C TS T A LGI + +L++ S +D+TI
Sbjct 25 LGTGSTVFYVLERIDNLLKSGKLKDVVCIPTSIDTELKARKLGIPLTTLEKHS-NIDITI 83
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D DE+ N + LIKGRGGAL+REK+V +S FI + DE K+ + G TGAV +E
Sbjct 84 DGTDEIDLN---LNLIKGRGGALVREKLVASSSSLFIIIGDESKLCT-NGLGMTGAVPIE 139
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
++ FG + + +L + TL G ++ + E+F++DN
Sbjct 140 ILTFGYEKIIENLLK--IYTLKG--------------------CTYKIRKRNGEIFITDN 177
Query 183 GNLCLDLFF 191
N +D FF
Sbjct 178 KNYIVDFFF 186
> ath:AT3G04790 ribose 5-phosphate isomerase-related; K01807 ribose
5-phosphate isomerase A [EC:5.3.1.6]
Length=276
Score = 99.8 bits (247), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 74/192 (38%), Positives = 106/192 (55%), Gaps = 30/192 (15%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
LGTGSTA+FAV+++ + + GEL DI TS+ T + A SLGI ++ LD + P +D+
Sbjct 68 LGTGSTAAFAVDQIGKLLSSGELYDIVGIPTSKRTEEQARSLGIPLVGLD--THPRIDLA 125
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG-AVA 120
ID ADEV N + L+KGRGGALLREK+VE + FI VAD+ K+V G +G A+
Sbjct 126 IDGADEVDPN---LDLVKGRGGALLREKMVEAVADKFIVVADDTKLVT--GLGGSGLAMP 180
Query 121 LEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVS 180
+EVVQF + ++ + G + R + K +V+
Sbjct 181 VEVVQFCWNFNLIRL--------------------QDLFKEFGCESKLRVDGDGKP-YVT 219
Query 181 DNGNLCLDLFFK 192
DN N +DL+FK
Sbjct 220 DNSNYIIDLYFK 231
> ath:AT2G01290 ribose-5-phosphate isomerase (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=265
Score = 98.6 bits (244), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 71/187 (37%), Positives = 105/187 (56%), Gaps = 26/187 (13%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGSTA AV+R+ E +RQG+L +I TS+ T++ A SLGI + LD + +D++I
Sbjct 56 LGTGSTAKHAVDRIGELLRQGKLENIVGIPTSKKTQEQALSLGIPLSDLDAHPV-IDLSI 114
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV + L+KGRGG+LLREK++E SK F+ + D+ K+V+ G+ A+ +E
Sbjct 115 DGADEV---DPFLNLVKGRGGSLLREKMIEGASKKFVVIVDDSKMVKHIG-GSKLALPVE 170
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
+V F + T + + S + E G A R E K FV+DN
Sbjct 171 IVPFCWKFTAEKLRSLL--------------------EGYGCEANLRLGEKGKA-FVTDN 209
Query 183 GNLCLDL 189
GN +D+
Sbjct 210 GNYIVDM 216
> ath:AT1G71100 RSW10; RSW10 (RADIAL SWELLING 10); ribose-5-phosphate
isomerase (EC:5.3.1.6); K01807 ribose 5-phosphate isomerase
A [EC:5.3.1.6]
Length=267
Score = 95.9 bits (237), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 62/131 (47%), Positives = 84/131 (64%), Gaps = 7/131 (5%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGSTA AV R++E +R+G+L DI TS T + A SLGI + LD + +D++I
Sbjct 52 LGTGSTAKHAVARISELLREGKLKDIIGIPTSTTTHEQAVSLGIPLSDLDSHPV-VDLSI 110
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG-AVAL 121
D ADEV + L+KGRGG+LLREK++E SK F+ + DE K+V+ G +G AV +
Sbjct 111 DGADEV---DPALNLVKGRGGSLLREKMIEGASKKFVVIVDESKLVK--YIGGSGLAVPV 165
Query 122 EVVQFGAQATR 132
EVV F TR
Sbjct 166 EVVPFCCDFTR 176
> dre:100332995 chloroplast ribose-5-phosphate isomerase-like
Length=291
Score = 95.5 bits (236), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 79/190 (41%), Positives = 101/190 (53%), Gaps = 31/190 (16%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
+GTGSTA V LAE++ G + TSE T +L LG+ + SLDE LP LD+T
Sbjct 85 IGTGSTAEEFVRLLAEKVAGG--LKVEGVPTSERTARLCVELGVPLKSLDE--LPELDLT 140
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVAL 121
ID ADEV + + LIKG GGALLREKIV S+ I +ADE KVV D G + +
Sbjct 141 IDGADEV---DASLRLIKGGGGALLREKIVACASQRMIVIADETKVV--DMLGAF-PLPI 194
Query 122 EVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSD 181
E+ FG ATR AV E+ A RLG+S + + LF++D
Sbjct 195 EINAFGQIATRIAV--------------------EKVASRLGLSGELKLRSSGDGLFMTD 234
Query 182 NGNLCLDLFF 191
G+L LD F
Sbjct 235 GGHLILDASF 244
> bbo:BBOV_III002460 17.m07239; ribose 5-phosphate isomerase A
family protein (EC:5.3.1.6); K01807 ribose 5-phosphate isomerase
A [EC:5.3.1.6]
Length=240
Score = 92.4 bits (228), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 66/189 (34%), Positives = 101/189 (53%), Gaps = 26/189 (13%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
LGTGSTAS+AV LA+++ +L D++ +TS AT +L LGI L D+ S+ +DV I
Sbjct 27 LGTGSTASYAVRYLAKQLSAAKLRDVTAVATSVATFRLMSELGIPSLEFDD-SVSIDVAI 85
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D AD + + LIKG GGAL REK+VE+ ++ I +AD K V+ V +E
Sbjct 86 DGADAI---DGDLNLIKGGGGALFREKLVELGARKVIIIADGTKKVKGHLL-QAFHVPVE 141
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
+VQFG T +S ++S +G V + + + L+++DN
Sbjct 142 IVQFGHVVT----ISRILSRVGPHVRSWTKRMNADGS-----------------LYLTDN 180
Query 183 GNLCLDLFF 191
N+ +D+ F
Sbjct 181 NNVIMDIEF 189
> tpv:TP03_0628 ribose 5-phosphate epimerase (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=363
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 64/193 (33%), Positives = 97/193 (50%), Gaps = 28/193 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEI---SLPLD 59
LGTG T++FA+ERLA I+ +T+I TS+AT A+ L I ++SLDE+ ++
Sbjct 30 LGTGRTSTFALERLANHIKTKSITNILGIPTSQATYLKAKELEIPLISLDEVIGSDRRIN 89
Query 60 VTIDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAV 119
+ ID AD V S + LIKG GGAL E +VE S I + DE K+ D + +
Sbjct 90 IAIDGADSV---DSDLNLIKGGGGALFTEFMVEQYSDQLIIMIDESKLCN-DHLLESHYL 145
Query 120 ALEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFV 179
+E+V+ T + + S S + V+ R + + LF+
Sbjct 146 PIEIVKGCHLTTCKQIYSKFKSQI--------------------VNWSVRMNSD-GSLFL 184
Query 180 SDNGNLCLDLFFK 192
+DNGN+ +D FK
Sbjct 185 TDNGNVIMDTQFK 197
> dre:407673 rpia, MGC103524, zgc:103524; ribose 5-phosphate isomerase
A (ribose 5-phosphate epimerase) (EC:5.3.1.6); K01807
ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=275
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 66/189 (34%), Positives = 89/189 (47%), Gaps = 28/189 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+G+GST +AV+RLAE++RQ +L +I C TS R+L G+ + LD LDV I
Sbjct 66 VGSGSTIVYAVDRLAEKVRQEKL-NIVCVPTSFQARQLILQHGLPLSDLDR-HPELDVAI 123
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV + + LIKG GG L +EKIV +K FI +AD K + V +E
Sbjct 124 DGADEV---DTALTLIKGGGGCLTQEKIVAGCAKHFIVIADYRKDSKALGQQWKKGVPVE 180
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
V+ V A+ GGE AV R ++ V+DN
Sbjct 181 VIPMAY----VPVSRAIARCFGGE-------------------AVLRMAVSKAGPVVTDN 217
Query 183 GNLCLDLFF 191
N LD F
Sbjct 218 SNFILDWKF 226
> xla:414499 rpia, MGC83218; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=235
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 70/191 (36%), Positives = 91/191 (47%), Gaps = 32/191 (16%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+G+GST AV RLAER++Q L + C TS R+L G+ + LD LDV I
Sbjct 26 IGSGSTIVHAVNRLAERVKQENLK-VVCVPTSFQARQLILQNGLVLTDLDR-HPELDVAI 83
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFG--TTGAVA 120
D ADEV S + LIKG GG L +EKIV +K+FI +AD K +FG V
Sbjct 84 DGADEV---DSDLNLIKGGGGCLAQEKIVAGCAKSFIVIADYRK--DSKNFGEQWKKGVP 138
Query 121 LEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVS 180
+EV+ V+ A+ S GG A R+ VS + V+
Sbjct 139 IEVIPM----AYVPVIKALQSRFGG-----------VAELRMAVS--------KAGPVVT 175
Query 181 DNGNLCLDLFF 191
DNGN LD F
Sbjct 176 DNGNFLLDWKF 186
> mmu:19895 Rpia, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=303
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 64/189 (33%), Positives = 90/189 (47%), Gaps = 28/189 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+G+GST AV+R+AER++Q L D+ C TS R+L G+ + LD+ +D+ I
Sbjct 95 IGSGSTIVHAVQRIAERVKQENL-DLICIPTSFQARQLILQYGLTLSDLDQ-HPEIDLAI 152
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV +++ LIKG GG L +EKIV + FI +AD K + + +E
Sbjct 153 DGADEV---DAELNLIKGGGGCLTQEKIVAGYASRFIVIADFRKDSKNLGDRWHKGIPIE 209
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
V+ V AV GGEV R+ V N+ V+DN
Sbjct 210 VIPM----AYVPVSRAVAQKFGGEV-----------ELRMAV--------NKAGPVVTDN 246
Query 183 GNLCLDLFF 191
GN LD F
Sbjct 247 GNFILDWKF 255
> cel:B0280.3 hypothetical protein; K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=251
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 64/192 (33%), Positives = 86/192 (44%), Gaps = 34/192 (17%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLP-LDVT 61
+G+GST + VE L + + G L DI C TS T++ G+ V LD S P LDV
Sbjct 38 VGSGSTVKYLVEYLKQGFQNGSLKDIICVPTSFLTKQWLIESGLPVSDLD--SHPELDVC 95
Query 62 IDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQP--DSFGTTGAV 119
ID ADEV Q IKG GG L +EKIV+ +K F +AD K + D + V
Sbjct 96 IDGADEV---DGQFTCIKGGGGCLAQEKIVQTAAKNFYVIADYLKDSKHLGDRYPN---V 149
Query 120 ALEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFV 179
+EV+ A Q +L ++ GG S R + V
Sbjct 150 PIEVLPLAA----QPLLRSIPRAEGG-------------------SCQLRQAVKKCGPIV 186
Query 180 SDNGNLCLDLFF 191
+DNGN +D F
Sbjct 187 TDNGNFIIDWQF 198
> hsa:22934 RPIA, RPI; ribose 5-phosphate isomerase A (EC:5.3.1.6);
K01807 ribose 5-phosphate isomerase A [EC:5.3.1.6]
Length=311
Score = 69.3 bits (168), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 58/189 (30%), Positives = 86/189 (45%), Gaps = 28/189 (14%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+G+GST AV+R+AER++Q L ++ C TS R+L G+ + LD +D+ I
Sbjct 102 IGSGSTIVHAVQRIAERVKQENL-NLVCIPTSFQARQLILQYGLTLSDLDR-HPEIDLAI 159
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADEV + + LIKG GG L +EKIV + FI +AD K + + +E
Sbjct 160 DGADEV---DADLNLIKGGGGCLTQEKIVAGYASRFIVIADFRKDSKNLGDQWHKGIPIE 216
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
V+ +AV +++ G R N+ V+DN
Sbjct 217 VIPMAYVPVSRAV-----------------------SQKFGGVVELRMAVNKAGPVVTDN 253
Query 183 GNLCLDLFF 191
GN LD F
Sbjct 254 GNFILDWKF 262
> eco:b2914 rpiA, ECK2910, JW5475, ygfC; ribose 5-phosphate isomerase,
constitutive (EC:5.3.1.6); K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=219
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 67/187 (35%), Positives = 94/187 (50%), Gaps = 41/187 (21%)
Query 3 LGTGSTASFAVERLAERIRQGELTDISCASTSEATRKLAESLGINVLSLDEISLPLDVTI 62
+GTGSTA+ ++ L Q E S+S+A+ + +SLGI+V L+E+ L + +
Sbjct 26 VGTGSTAAHFIDALGTMKGQIE----GAVSSSDASTEKLKSLGIHVFDLNEVD-SLGIYV 80
Query 63 DFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTGAVALE 122
D ADE+ NG M +IKG G AL REKI+ ++ FIC+AD K Q D G + +E
Sbjct 81 DGADEI--NG-HMQMIKGGGAALTREKIIASVAEKFICIADASK--QVDILGKF-PLPVE 134
Query 123 VVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVSAVFRHHENRKELFVSDN 182
V+ R AV +V LGG PE R GV V+DN
Sbjct 135 VIPM----ARSAVARQLVK-LGGR--------PE---YRQGV--------------VTDN 164
Query 183 GNLCLDL 189
GN+ LD+
Sbjct 165 GNVILDV 171
> sce:YOR095C RKI1; Ribose-5-phosphate ketol-isomerase, catalyzes
the interconversion of ribose 5-phosphate and ribulose 5-phosphate
in the pentose phosphate pathway; participates in
pyridoxine biosynthesis (EC:5.3.1.6); K01807 ribose 5-phosphate
isomerase A [EC:5.3.1.6]
Length=258
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 65/128 (50%), Gaps = 11/128 (8%)
Query 3 LGTGSTASFAVERLAERIRQGELTDIS----CASTSEATRKLAESLGINVLSLDEISLPL 58
+G+GST + ER+ + + + +++ C T +R L + + S+++ +
Sbjct 44 IGSGSTVVYVAERIGQYLHDPKFYEVASKFICIPTGFQSRNLILDNKLQLGSIEQYPR-I 102
Query 59 DVTIDFADEVLKNGSQMVLIKGRGGALLREKIVEVNSKAFICVADEGKVVQPDSFGTTG- 117
D+ D ADEV +N + LIKG G L +EK+V ++K FI VAD K P G
Sbjct 103 DIAFDGADEVDEN---LQLIKGGGACLFQEKLVSTSAKTFIVVADSRK-KSPKHLGKNWR 158
Query 118 -AVALEVV 124
V +E+V
Sbjct 159 QGVPIEIV 166
> tgo:TGME49_030950 hypothetical protein
Length=2170
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 27/80 (33%), Positives = 38/80 (47%), Gaps = 6/80 (7%)
Query 108 VQP--DSFGTTGAVALEVVQFGAQATRQAVLSAVVSTLGGEVLEAEEETPEEAAERLGVS 165
QP D+ G+T AV EV+ RQ + GG AE E E+A R ++
Sbjct 78 TQPRHDARGSTHAVLEEVIALLNDVRRQG--GRAIWPCGGAGDTAETEQGEQALFRRDLA 135
Query 166 AVFRHHENR--KELFVSDNG 183
++FRH R + F+SD G
Sbjct 136 SLFRHSLRRCSRRAFLSDRG 155
Lambda K H
0.315 0.131 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5558593832
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40