bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2195_orf1
Length=144
Score E
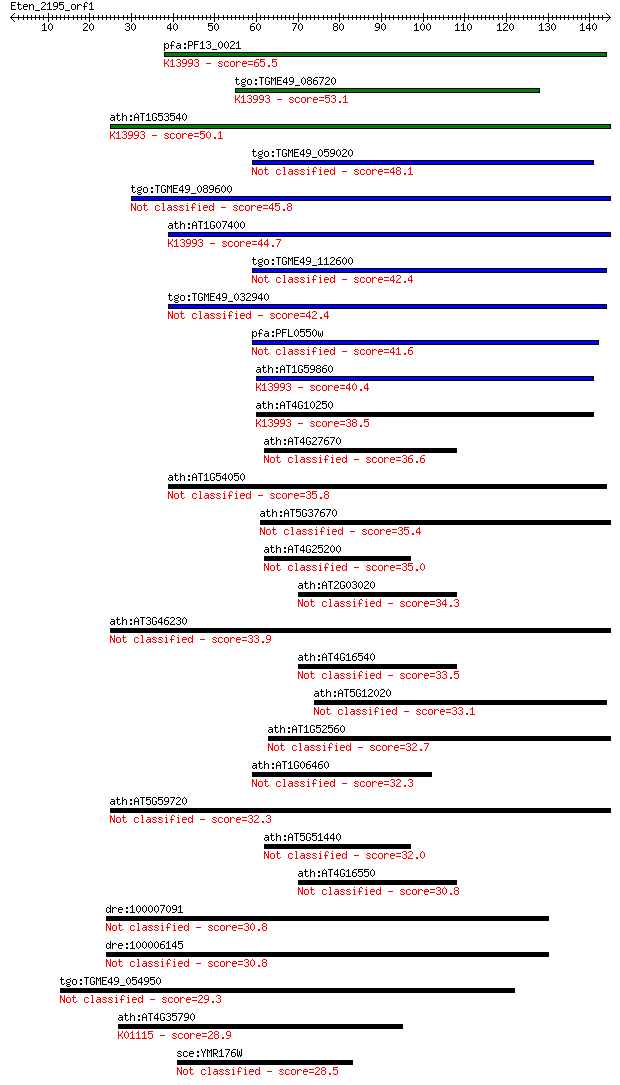
Sequences producing significant alignments: (Bits) Value
pfa:PF13_0021 small heat shock protein, putative; K13993 HSP20... 65.5 6e-11
tgo:TGME49_086720 heat shock protein 28 ; K13993 HSP20 family ... 53.1 3e-07
ath:AT1G53540 17.6 kDa class I small heat shock protein (HSP17... 50.1 2e-06
tgo:TGME49_059020 bradyzoite antigen, putative 48.1 9e-06
tgo:TGME49_089600 Hsp20/alpha crystallin domain-containing pro... 45.8 4e-05
ath:AT1G07400 17.8 kDa class I heat shock protein (HSP17.8-CI)... 44.7 1e-04
tgo:TGME49_112600 small heat shock protein 21 (EC:1.1.2.4) 42.4 5e-04
tgo:TGME49_032940 small heat shock protein 20 42.4
pfa:PFL0550w HSP20-like chaperone 41.6 0.001
ath:AT1G59860 17.6 kDa class I heat shock protein (HSP17.6A-CI... 40.4 0.002
ath:AT4G10250 ATHSP22.0; K13993 HSP20 family protein 38.5 0.007
ath:AT4G27670 HSP21; HSP21 (HEAT SHOCK PROTEIN 21) 36.6 0.029
ath:AT1G54050 17.4 kDa class III heat shock protein (HSP17.4-C... 35.8 0.045
ath:AT5G37670 15.7 kDa class I-related small heat shock protei... 35.4 0.063
ath:AT4G25200 ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL HE... 35.0 0.082
ath:AT2G03020 heat shock protein-related 34.3 0.14
ath:AT3G46230 ATHSP17.4 33.9 0.18
ath:AT4G16540 heat shock protein-related 33.5 0.24
ath:AT5G12020 HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN) 33.1 0.33
ath:AT1G52560 26.5 kDa class I small heat shock protein-like (... 32.7 0.43
ath:AT1G06460 ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 32.1) 32.3 0.46
ath:AT5G59720 HSP18.2 (heat shock protein 18.2) 32.3 0.59
ath:AT5G51440 23.5 kDa mitochondrial small heat shock protein ... 32.0 0.73
ath:AT4G16550 heat shock protein-related 30.8 1.5
dre:100007091 zinc finger protein-like 30.8 1.6
dre:100006145 zinc finger protein-like 30.8 1.6
tgo:TGME49_054950 hypothetical protein 29.3 4.9
ath:AT4G35790 ATPLDDELTA; phospholipase D (EC:3.1.4.4); K01115... 28.9 5.8
sce:YMR176W ECM5; Ecm5p 28.5 8.5
> pfa:PF13_0021 small heat shock protein, putative; K13993 HSP20
family protein
Length=211
Score = 65.5 bits (158), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 35/106 (33%), Positives = 59/106 (55%), Gaps = 14/106 (13%)
Query 38 LLQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRG 97
LL P SS + +S+ +P +D+ D +L + D+PGL+K+D+++ +G+L I G
Sbjct 86 LLSKPFSSMFRRDGYSN---VPAMDVLDKEKHLEIKMDVPGLNKEDVQINLDDGKLEISG 142
Query 98 HSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEA 143
S +Q+ +QQ R++++ERC SSF+RS LP
Sbjct 143 EFKKS-------HEQKDEQQ----RYYIKERCESSFYRSFTLPENV 177
> tgo:TGME49_086720 heat shock protein 28 ; K13993 HSP20 family
protein
Length=276
Score = 53.1 bits (126), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/73 (31%), Positives = 48/73 (65%), Gaps = 13/73 (17%)
Query 55 SSCIPRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQ 114
+ +P++D++DTG ++HAD+PG+D+++++V+ +G L I G Q+ +
Sbjct 166 AGSMPKVDMKDTGSEFVVHADVPGMDRENLRVDVHDGVLRISG------------TQRDE 213
Query 115 QQQQQDGRWFVQE 127
+QQQ++G +++QE
Sbjct 214 KQQQEEG-FYLQE 225
> ath:AT1G53540 17.6 kDa class I small heat shock protein (HSP17.6C-CI)
(AA 1-156); K13993 HSP20 family protein
Length=157
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 62/121 (51%), Gaps = 26/121 (21%)
Query 25 QGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKKDI 84
+G L+P SGL PA ++ F+++ ++D R+T + + ADLPGL K+++
Sbjct 28 EGFLTP-----SGLANAPAMDVAA---FTNA----KVDWRETPEAHVFKADLPGLRKEEV 75
Query 85 KVETQNGRLI-IRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEA 143
KVE ++G ++ I G S+ + ++++ +W ER S F R LP A
Sbjct 76 KVEVEDGNILQISGERSN-------------ENEEKNDKWHRVERSSGKFTRRFRLPENA 122
Query 144 Q 144
+
Sbjct 123 K 123
> tgo:TGME49_059020 bradyzoite antigen, putative
Length=229
Score = 48.1 bits (113), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 43/86 (50%), Gaps = 15/86 (17%)
Query 59 PRLDIR--DTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQ 116
PR+D+ +++ ADLPGL K D+ +E NG ++I+G +S ++ +
Sbjct 119 PRVDVEFDSKKKEMIILADLPGLQKDDVTIEVDNGAIVIKGEKTS-----------KEAE 167
Query 117 QQQDGRW--FVQERCSSSFFRSLPLP 140
+ DG+ + ER S F R LP
Sbjct 168 KVDDGKTKNILTERVSGYFARRFQLP 193
> tgo:TGME49_089600 Hsp20/alpha crystallin domain-containing protein
Length=272
Score = 45.8 bits (107), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 34/117 (29%), Positives = 52/117 (44%), Gaps = 15/117 (12%)
Query 30 PWSSSSSGLLQWPASSSSSSNWFSHSSCIPRLD-IRDTGDN-LLLHADLPGLDKKDIKVE 87
P ++ S + + S+ N S PRLD D N L+L DLPG +KKD+++E
Sbjct 136 PGVAAPSTIFEVGPGSAEICNKISFR---PRLDAYYDANRNRLVLLFDLPGFEKKDVEIE 192
Query 88 TQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEAQ 144
G L I G + ++ + QD ++ER F+R LP A+
Sbjct 193 LDKGALAISGERP----------KLEESKLGQDCNNIIKERSFGFFYRKFQLPGNAE 239
> ath:AT1G07400 17.8 kDa class I heat shock protein (HSP17.8-CI);
K13993 HSP20 family protein
Length=157
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 56/106 (52%), Gaps = 16/106 (15%)
Query 39 LQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGH 98
LQ+P+S S ++ +++ R+D ++T + + ADLPG+ K+++KVE ++ ++
Sbjct 32 LQFPSSLSGETSAITNA----RVDWKETAEAHVFKADLPGMKKEEVKVEIEDDSVL---- 83
Query 99 SSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEAQ 144
S ++ +++++QD W ER S F R LP +
Sbjct 84 -------KISGERHVEKEEKQDT-WHRVERSSGQFSRKFKLPENVK 121
> tgo:TGME49_112600 small heat shock protein 21 (EC:1.1.2.4)
Length=195
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 11/87 (12%)
Query 59 PRLD-IRDTGDN-LLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQ 116
PRLD DT + +++ DLPG +KKDI VE + +II G + + +
Sbjct 83 PRLDAYYDTAAHKIVMLFDLPGFEKKDISVEVTDHAIIISG---------TRDRLNDKVL 133
Query 117 QQQDGRWFVQERCSSSFFRSLPLPPEA 143
+ R ++ER F R LP A
Sbjct 134 YGEKSRELIKERAFGHFCRKFQLPTNA 160
> tgo:TGME49_032940 small heat shock protein 20
Length=192
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 50/111 (45%), Gaps = 25/111 (22%)
Query 39 LQWPASSSSSSNWFSHSSCIPRLDI----RDTGDNLLLHADLPGLDKKDIKVETQNGRLI 94
+Q PA S W P +DI +++ +L++ DLPG K D+ VE G+L
Sbjct 56 IQAPAEIRSKITWR------PGVDIFFDKKESSIDLVM--DLPGFTKDDVSVEVGEGQLF 107
Query 95 IRGHSSSSSSSSSSPQQQQQQQQQQDGRWF--VQERCSSSFFRSLPLPPEA 143
I G P+ + + +++ + ER + FFRS LPP A
Sbjct 108 ISG-----------PRSKNELREKYGANLVLSIHERPTGFFFRSFQLPPNA 147
> pfa:PFL0550w HSP20-like chaperone
Length=231
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/83 (22%), Positives = 41/83 (49%), Gaps = 7/83 (8%)
Query 59 PRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQ 118
P+++I T D +L D+PG+ K+++KVE + G L + G + P ++ +++
Sbjct 119 PKIEIYSTCDFAVLMMDIPGVSKENLKVELEKGLLKVYG-------NKYKPHIEELEKRN 171
Query 119 QDGRWFVQERCSSSFFRSLPLPP 141
+ ++ F + +PP
Sbjct 172 EYHTKIIERLNEYYFCKIFQMPP 194
> ath:AT1G59860 17.6 kDa class I heat shock protein (HSP17.6A-CI);
K13993 HSP20 family protein
Length=155
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 43/81 (53%), Gaps = 12/81 (14%)
Query 60 RLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQ 119
R+D ++T + + ADLPG+ K+++KVE ++ ++ S ++ +++++Q
Sbjct 47 RVDWKETAEAHVFKADLPGMKKEEVKVEIEDDSVL-----------KISGERHVEKEEKQ 95
Query 120 DGRWFVQERCSSSFFRSLPLP 140
D W ER S F R LP
Sbjct 96 DT-WHRVERSSGGFSRKFRLP 115
> ath:AT4G10250 ATHSP22.0; K13993 HSP20 family protein
Length=195
Score = 38.5 bits (88), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/82 (25%), Positives = 43/82 (52%), Gaps = 14/82 (17%)
Query 60 RLDIRDTGDNLLLHADLPGLDKKDIKVET-QNGRLIIRGHSSSSSSSSSSPQQQQQQQQQ 118
R+D ++T + + D+PGL K ++K+E +NG L + G ++++++++
Sbjct 72 RVDWKETAEGHEIMLDIPGLKKDEVKIEVEENGVLRVSG-------------ERKREEEK 118
Query 119 QDGRWFVQERCSSSFFRSLPLP 140
+ +W ER F+R LP
Sbjct 119 KGDQWHRVERSYGKFWRQFKLP 140
> ath:AT4G27670 HSP21; HSP21 (HEAT SHOCK PROTEIN 21)
Length=227
Score = 36.6 bits (83), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 62 DIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSS 107
DI++ + + D+PGL K+D+K+ ++ L+I+G S S
Sbjct 130 DIKEEEHEIKMRFDMPGLSKEDVKISVEDNVLVIKGEQKKEDSDDS 175
> ath:AT1G54050 17.4 kDa class III heat shock protein (HSP17.4-CIII)
Length=155
Score = 35.8 bits (81), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 28/112 (25%), Positives = 55/112 (49%), Gaps = 18/112 (16%)
Query 39 LQWPASSSSSSNWFSH----SSCIPRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGR-L 93
L P S S SN S S+ IP +DI ++ + + D+PG+ K DI+V + R L
Sbjct 19 LILPISRSGESNNESRGRGSSNNIP-IDILESPKEYIFYLDIPGISKSDIQVTVEEERTL 77
Query 94 IIRGHSSSSSSSSSSPQQQQQQQQQQDGRWFV--QERCSSSFFRSLPLPPEA 143
+I+ S+ ++++ + ++G ++ + R + + + LP +A
Sbjct 78 VIK----------SNGKRKRDDDESEEGSKYIRLERRLAQNLVKKFRLPEDA 119
> ath:AT5G37670 15.7 kDa class I-related small heat shock protein-like
(HSP15.7-CI)
Length=137
Score = 35.4 bits (80), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 21/90 (23%), Positives = 46/90 (51%), Gaps = 19/90 (21%)
Query 61 LDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLI-IRGHSSSSSSSSSSPQQQQQQQQQQ 119
+D ++ ++ + ++PG +K+DIKV+ + G ++ IRG + ++++++
Sbjct 24 IDWMESNNSHIFKINVPGYNKEDIKVQIEEGNVLSIRG-------------EGIKEEKKE 70
Query 120 DGRWFVQERCS-----SSFFRSLPLPPEAQ 144
+ W V ER + S F R + LP +
Sbjct 71 NLVWHVAEREAFSGGGSEFLRRIELPENVK 100
> ath:AT4G25200 ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL
HEAT SHOCK PROTEIN 23.6)
Length=210
Score = 35.0 bits (79), Expect = 0.082, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 62 DIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIR 96
DI++ D L L D+PGL ++D+K+ + L+IR
Sbjct 110 DIKEKDDALYLRIDMPGLSREDVKLALEQDTLVIR 144
> ath:AT2G03020 heat shock protein-related
Length=255
Score = 34.3 bits (77), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 70 LLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSS 107
L + AD+PG+ K++ V NGR+ + G + + S SS
Sbjct 180 LFVRADMPGVPKENFTVSVTNGRVKVTGQAPAVSHDSS 217
> ath:AT3G46230 ATHSP17.4
Length=156
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 59/121 (48%), Gaps = 27/121 (22%)
Query 25 QGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKKDI 84
+G L+P GL PA ++ F+++ ++D R+T + + AD+PGL K+++
Sbjct 28 EGFLTP------GLTNAPAKDVAA---FTNA----KVDWRETPEAHVFKADVPGLKKEEV 74
Query 85 KVETQNGRLI-IRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEA 143
KVE ++G ++ I G SS + +++ W ER S F R LP A
Sbjct 75 KVEVEDGNILQISGERSS-------------ENEEKSDTWHRVERSSGKFMRRFRLPENA 121
Query 144 Q 144
+
Sbjct 122 K 122
> ath:AT4G16540 heat shock protein-related
Length=232
Score = 33.5 bits (75), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 70 LLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSS 107
L + AD+PG+ K++ V NGR+ + G + + S SS
Sbjct 157 LFVRADMPGVPKENFTVSVTNGRVKVTGEAPALSHDSS 194
> ath:AT5G12020 HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN)
Length=155
Score = 33.1 bits (74), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 16/70 (22%), Positives = 33/70 (47%), Gaps = 11/70 (15%)
Query 74 ADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSF 133
D+PG+ +IKV+ +N +++ S ++Q++ ++ + ++ ER F
Sbjct 60 VDMPGIKGDEIKVQVENDNVLV-----------VSGERQRENKENEGVKYVRMERRMGKF 108
Query 134 FRSLPLPPEA 143
R LP A
Sbjct 109 MRKFQLPENA 118
> ath:AT1G52560 26.5 kDa class I small heat shock protein-like
(HSP26.5-P)
Length=225
Score = 32.7 bits (73), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/82 (23%), Positives = 40/82 (48%), Gaps = 11/82 (13%)
Query 63 IRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDGR 122
+++ D L ++PGL K+D+K+ +G L I+G + ++++ ++D
Sbjct 122 VKEQDDCYKLRYEVPGLTKEDVKITVNDGILTIKGDHKA---------EEEKGSPEEDEY 172
Query 123 WFVQERCSSSFFRSLPLPPEAQ 144
W + + SL LP +A+
Sbjct 173 W--SSKSYGYYNTSLSLPDDAK 192
> ath:AT1G06460 ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 32.1)
Length=285
Score = 32.3 bits (72), Expect = 0.46, Method: Compositional matrix adjust.
Identities = 13/43 (30%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 59 PRLDIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIRGHSSS 101
PR ++ ++ + ++ +LPG DI+VE N L + G +S
Sbjct 186 PRSNVAESTHSYVVAIELPGASINDIRVEVDNTNLTVTGRRTS 228
> ath:AT5G59720 HSP18.2 (heat shock protein 18.2)
Length=161
Score = 32.3 bits (72), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 60/121 (49%), Gaps = 24/121 (19%)
Query 25 QGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKKDI 84
+G +P SS L AS++ F+++ R+D ++T + + ADLPGL K+++
Sbjct 28 EGFFTP----SSALAN--ASTARDVAAFTNA----RVDWKETPEAHVFKADLPGLKKEEV 77
Query 85 KVETQNGRLI-IRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERCSSSFFRSLPLPPEA 143
KVE ++ ++ I G S ++ ++++ +W ER S F R LP A
Sbjct 78 KVEVEDKNVLQISGERS-------------KENEEKNDKWHRVERASGKFMRRFRLPENA 124
Query 144 Q 144
+
Sbjct 125 K 125
> ath:AT5G51440 23.5 kDa mitochondrial small heat shock protein
(HSP23.5-M)
Length=210
Score = 32.0 bits (71), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 62 DIRDTGDNLLLHADLPGLDKKDIKVETQNGRLIIR 96
++++ D L L D+PGL ++D+K+ + L+IR
Sbjct 112 NVKEKDDALHLRIDMPGLSREDVKLALEQNTLVIR 146
> ath:AT4G16550 heat shock protein-related
Length=743
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 70 LLLHADLPGLDKKDIKVETQNGRLIIRGHSSSSSSSSS 107
L + D+PG+ K++ V NGR+ + G + + S SS
Sbjct 668 LYVRVDMPGVPKENFTVAVMNGRVRVTGEAPAISHDSS 705
> dre:100007091 zinc finger protein-like
Length=666
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 43/111 (38%), Gaps = 17/111 (15%)
Query 24 QQGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIP-----RLDIRDTGDNLLLHADLPG 78
QG + + L P +S+ S FS +S + RL L + LP
Sbjct 560 NQGRFPDLAQMARRYLSAPCTSTDSERLFSAASHVIDEKRNRLSCEKAEKLLFIKKILPH 619
Query 79 LDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERC 129
KKD K H+SSS + +++ +++ Q W+ + +C
Sbjct 620 FLKKDTK------------HASSSPRPTVDSKKEVEEEAQHTALWYTRVKC 658
> dre:100006145 zinc finger protein-like
Length=666
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 25/111 (22%), Positives = 43/111 (38%), Gaps = 17/111 (15%)
Query 24 QQGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIP-----RLDIRDTGDNLLLHADLPG 78
QG + + L P +S+ S FS +S + RL L + LP
Sbjct 560 NQGRFPDLAQMARRYLSAPCTSTDSERLFSAASHVIDEKRNRLSCEKAEKLLFIKKILPH 619
Query 79 LDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDGRWFVQERC 129
KKD K H+SSS + +++ +++ Q W+ + +C
Sbjct 620 FLKKDTK------------HASSSPRPTVDSKKEVEEEAQHTALWYTRVKC 658
> tgo:TGME49_054950 hypothetical protein
Length=1869
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 30/114 (26%), Positives = 47/114 (41%), Gaps = 6/114 (5%)
Query 13 QLAGAGPAGAQQQGLLSPWSSSSSGLLQWPASSSSSSNWFSHSSCIPRLDIRDTGDNLLL 72
+L G PA + QGLLS S++ A S +N C P ++ R+ N
Sbjct 1535 RLGGLTPA-SPAQGLLSVDCQSTTCACSSVARPRSCTNGLCRDVCGPPVETREFRPNAPG 1593
Query 73 HADLPG-----LDKKDIKVETQNGRLIIRGHSSSSSSSSSSPQQQQQQQQQQDG 121
A LP LD V G + RG S SS + +++ ++ ++ G
Sbjct 1594 AAPLPAFAWFSLDPHLRAVREAEGDSLCRGGDSPSSRGGDAEKKKSERVKETAG 1647
> ath:AT4G35790 ATPLDDELTA; phospholipase D (EC:3.1.4.4); K01115
phospholipase D [EC:3.1.4.4]
Length=868
Score = 28.9 bits (63), Expect = 5.8, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 38/76 (50%), Gaps = 8/76 (10%)
Query 27 LLSP-WSSSSSGLLQWPASSSSS-------SNWFSHSSCIPRLDIRDTGDNLLLHADLPG 78
+LSP ++SS GL + AS SSS F+H +D + G+N + A + G
Sbjct 337 VLSPRYASSKLGLFKQQASPSSSIYIMTVVGTLFTHHQKCVLVDTQAVGNNRKVTAFIGG 396
Query 79 LDKKDIKVETQNGRLI 94
LD D + +T R++
Sbjct 397 LDLCDGRYDTPEHRIL 412
> sce:YMR176W ECM5; Ecm5p
Length=1411
Score = 28.5 bits (62), Expect = 8.5, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 41 WPASSSSSSNWFSHSSCIPRLDIRDTGDNLLLHADLPGLDKK 82
W SS ++ W S +P LD+ N LL+++ P L KK
Sbjct 683 WLRFSSEAAKWTSKMGFLPGLDVNQLLINALLNSNNPVLRKK 724
Lambda K H
0.310 0.123 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2814663556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40