bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2140_orf2
Length=241
Score E
Sequences producing significant alignments: (Bits) Value
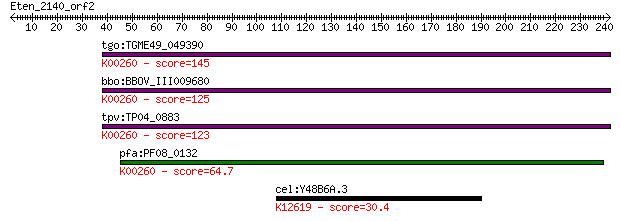
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 145 2e-34
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 125 2e-28
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 123 5e-28
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 64.7 3e-10
cel:Y48B6A.3 xrn-2; XRN (mouse/S. cerevisiae) ribonuclease rel... 30.4 5.2
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 145 bits (365), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 90/209 (43%), Positives = 123/209 (58%), Gaps = 31/209 (14%)
Query 38 MSEKYGAQFAQVQQLLLQKGWFAEPVATEHLLDYYNHLGLNEFYFKAASPKSIADNLLAV 97
MS+KY AQ+ +V+Q+L Q+G ++ V + + YYNHLGLNE+YF+ A+ +++A NL V
Sbjct 93 MSQKYAAQYEEVKQILAQRGDLSKRVISANADAYYNHLGLNEYYFQTATAQTVASNLACV 152
Query 98 IAARALHSVSGDDYFPEIRQRDERSGDVFALAKASLTASGAPRNYLVERAFELQFLDMGW 157
IAA+ L+ S DYFP+I+Q ER G+VF LA+ASL A +N+ VER E +FL +
Sbjct 153 IAAKILNEFSHTDYFPQIQQ--ERDGEVFLLARASLRNRKASQNFQVEREIEKKFLGLS- 209
Query 158 GLRAPPQQQQRQQQQQRQQQRRRKQQMRMQCYRSTGSVFGMDGGA---ERLRTYFFQVAK 214
G P RMQCYRS+GS F D A ERLRTYF Q +
Sbjct 210 GKTTP---------------------YRMQCYRSSGSFF--DDPADENERLRTYFLQQPE 246
Query 215 FPKRPEQ--LSPKEKDLSLLLDTDFYASK 241
+ + + P E DLS LLD F+A+K
Sbjct 247 YCAETTKDGVDPSETDLSKLLDVYFFANK 275
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 125 bits (313), Expect = 2e-28, Method: Composition-based stats.
Identities = 79/208 (37%), Positives = 110/208 (52%), Gaps = 32/208 (15%)
Query 38 MSEKYGAQFAQVQQLLLQKGWFAEPVATEHLLDYYNHLGLNEFYFKAASPKSIADNLLAV 97
+S++Y QF +V +++ F++ + + YYN LGLNE+YF ++ K IA+N+++V
Sbjct 30 LSDRYATQFQEVYKIVKDLKTFSDDTINQTMDQYYNKLGLNEYYFSTSNAKMIANNMVSV 89
Query 98 IAARALHSVSGDDYFPEIRQRDERSGDVFALAKASLTASGAPRNYLVERAFELQFLDMGW 157
+ A+ LH SG DYFP I Q + G VF + +ASL + Y VE+ E +FL G
Sbjct 90 LTAKILHENSGSDYFPLIEQVHD--GRVFIITRASLLNHKLSQIYAVEKTVEQRFLHFGD 147
Query 158 GLRAPPQQQQRQQQQQRQQQRRRKQQMRMQCYRSTGSVFGMDGGA---ERLRTYFFQVAK 214
R RMQC+RST S+F D A ERLR YFFQ+
Sbjct 148 VTRP---------------------VWRMQCFRSTNSMF--DDPANLFERLRIYFFQLPN 184
Query 215 FPK-RPEQLSPKEKDLSLLLDTDFYASK 241
F + PE P E LS LLD DFY +K
Sbjct 185 FAENNPE---PGELRLSKLLDKDFYKNK 209
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 123 bits (309), Expect = 5e-28, Method: Composition-based stats.
Identities = 73/206 (35%), Positives = 113/206 (54%), Gaps = 28/206 (13%)
Query 38 MSEKYGAQFAQVQQLLLQKGWFAEPVATEHLLDYYNHLGLNEFYFKAASPKSIADNLLAV 97
+S++Y QF++V +L+ F++ E + DYYN LGLNE+YF ++ IA+N+++V
Sbjct 29 LSDRYCVQFSEVSELIRSMKTFSDDKIIEIMHDYYNELGLNEYYFSSSPTALIANNVISV 88
Query 98 IAARALHSVSGDDYFPEIRQRDERSGDVFALAKASLTASGAPRNYLVERAFELQFLDMGW 157
++A+ LH SG ++FP I Q E S F +A+ S + NYLVE+ E ++ ++
Sbjct 89 MSAKVLHENSGSNFFPTIEQITEDSA--FIIARTSSVNRKSSENYLVEQRVEGKYFNL-- 144
Query 158 GLRAPPQQQQRQQQQQRQQQRRRKQQMRMQCYRSTGSVFG-MDGGAERLRTYFFQVAKFP 216
RK RMQC+RS+GSVF + +RL+TYF Q KF
Sbjct 145 -------------------DDDRKTCWRMQCFRSSGSVFKETEHIFDRLKTYFLQKPKFT 185
Query 217 -KRPEQLSPKEKDLSLLLDTDFYASK 241
P+ P E +L+ LLD +FY +K
Sbjct 186 VNNPD---PGETELANLLDREFYLNK 208
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 64.7 bits (156), Expect = 3e-10, Method: Composition-based stats.
Identities = 48/198 (24%), Positives = 85/198 (42%), Gaps = 32/198 (16%)
Query 45 QFAQVQQLLLQKGWFAEPVATEHLLDYYNHLGLNEFYFKAASPKSIADNLLAVIAARALH 104
++ Q ++LL F++ + + Y+N LG N+F+F+ SP+ I+ ++ +I A+
Sbjct 35 KYEQTKELLKSYNLFSDHLINYSIDFYFNKLGFNKFHFEETSPELISKVVVCIITAKINE 94
Query 105 SVSGDDYFPEIRQRDERSGDVFALAKASLTASGAPRNYLVERAFE---LQFLDMGWGLRA 161
S D YFP + + +F + + + NY +E+ E F DM
Sbjct 95 QYSSDKYFPTFEETHDNV--IFIITRVFADDNKTRLNYKMEKKIEEKYFNFSDMS----- 147
Query 162 PPQQQQRQQQQQRQQQRRRKQQMRMQCYRSTGSVFGMDGG-AERLRTYFFQVAKFPKRPE 220
K R++ +RS SVF + E LRTY ++ + +
Sbjct 148 -------------------KDCYRLKSFRSVHSVFDKEHTYQEPLRTYILELPTY--NDD 186
Query 221 QLSPKEKDLSLLLDTDFY 238
+ E DL L+D +FY
Sbjct 187 IIKENETDLKKLMDVNFY 204
> cel:Y48B6A.3 xrn-2; XRN (mouse/S. cerevisiae) ribonuclease related
family member (xrn-2); K12619 5'-3' exoribonuclease 2
[EC:3.1.13.-]
Length=975
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 41/86 (47%), Gaps = 4/86 (4%)
Query 108 GDDYFPEIRQRDERSGDVFALAKASLTASGAPRNYLVERAF-ELQFLDM---GWGLRAPP 163
G+D+ P + + R G + L K + YL + EL ++M G G
Sbjct 342 GNDFLPHLPSLEIREGAIDRLIKLYKEMVYQMKGYLTKDGIPELDRVEMIMKGLGRVEDE 401
Query 164 QQQQRQQQQQRQQQRRRKQQMRMQCY 189
++RQQ ++R Q+ +R ++ RMQ Y
Sbjct 402 IFKRRQQDEERFQENQRNKKARMQMY 427
Lambda K H
0.320 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8466654768
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40