bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2128_orf1
Length=91
Score E
Sequences producing significant alignments: (Bits) Value
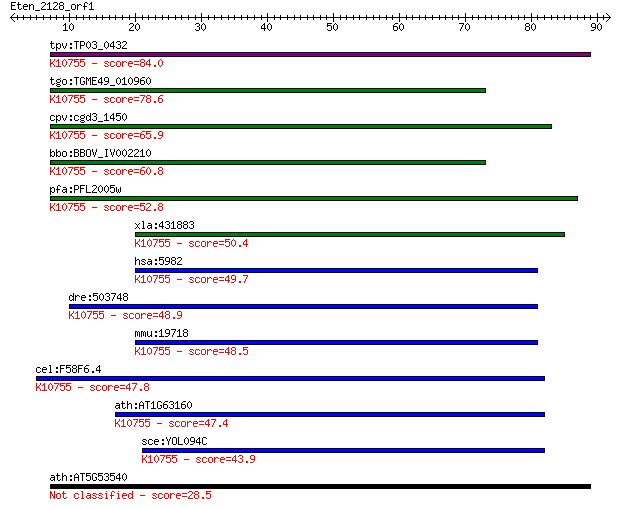
tpv:TP03_0432 replication factor C subunit 4; K10755 replicati... 84.0 1e-16
tgo:TGME49_010960 replication factor C subunit, putative (EC:2... 78.6 5e-15
cpv:cgd3_1450 replication factor C like AAA+ ATpase ; K10755 r... 65.9 3e-11
bbo:BBOV_IV002210 21.m03053; replication factor C subunit 4; K... 60.8 1e-09
pfa:PFL2005w replication factor C subunit 4; K10755 replicatio... 52.8 3e-07
xla:431883 rfc2, MGC81391, rfc40; replication factor C (activa... 50.4 1e-06
hsa:5982 RFC2, A1, MGC3665, RFC40; replication factor C (activ... 49.7 3e-06
dre:503748 rfc2, zgc:110810; replication factor C (activator 1... 48.9 4e-06
mmu:19718 Rfc2, 2610008M13Rik, 40kDa, AI326953, MGC117486, Rec... 48.5 5e-06
cel:F58F6.4 rfc-2; RFC (DNA replication factor) family member ... 47.8 7e-06
ath:AT1G63160 replication factor C 40 kDa, putative; K10755 re... 47.4 1e-05
sce:YOL094C RFC4; Rfc4p; K10755 replication factor C subunit 2/4 43.9
ath:AT5G53540 MSP1 protein, putative / intramitochondrial sort... 28.5 5.6
> tpv:TP03_0432 replication factor C subunit 4; K10755 replication
factor C subunit 2/4
Length=324
Score = 84.0 bits (206), Expect = 1e-16, Method: Composition-based stats.
Identities = 37/82 (45%), Positives = 57/82 (69%), Gaps = 2/82 (2%)
Query 7 WYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLS 66
W A +L LG++P+D++ T R VL+ D+ EH+LLE++ V ++HMTM GLS
Sbjct 245 WRQAHEKVDQLLELGHSPVDILVTMRNVLKTMDAP--EHVLLEYIKSVALSHMTMVNGLS 302
Query 67 TELQMEKMLAQLCKVAISFRPS 88
T+LQ+EK+LA LCK+A++ R +
Sbjct 303 TQLQLEKLLANLCKIALALRTT 324
> tgo:TGME49_010960 replication factor C subunit, putative (EC:2.7.7.7);
K10755 replication factor C subunit 2/4
Length=336
Score = 78.6 bits (192), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 36/66 (54%), Positives = 47/66 (71%), Gaps = 0/66 (0%)
Query 7 WYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLS 66
W A +A +L GYTP+DVV T R VL R ++E +EH+LLE+L VG+ HMTM+ GLS
Sbjct 271 WREAHDIAAELLRRGYTPMDVVLTTRSVLSRFENECKEHILLEYLKYVGLAHMTMSAGLS 330
Query 67 TELQME 72
T LQ+E
Sbjct 331 TPLQLE 336
> cpv:cgd3_1450 replication factor C like AAA+ ATpase ; K10755
replication factor C subunit 2/4
Length=339
Score = 65.9 bits (159), Expect = 3e-11, Method: Composition-based stats.
Identities = 32/76 (42%), Positives = 46/76 (60%), Gaps = 2/76 (2%)
Query 7 WYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLS 66
W A S+ + GY+PLD+V T R VL+R +L E +LE+L VG H M G +
Sbjct 255 WRLAHSIVEELFIGGYSPLDIVITMRNVLKR--YQLPERAILEYLKEVGRCHFVMLDGCA 312
Query 67 TELQMEKMLAQLCKVA 82
T LQ++K+L QLC ++
Sbjct 313 TPLQLDKLLGQLCMIS 328
> bbo:BBOV_IV002210 21.m03053; replication factor C subunit 4;
K10755 replication factor C subunit 2/4
Length=306
Score = 60.8 bits (146), Expect = 1e-09, Method: Composition-based stats.
Identities = 28/66 (42%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Query 7 WYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLS 66
W A A +L+LG++P D++ T R VL+ + EH+L EFL + ++HMTM GLS
Sbjct 243 WRPAHEKAEDLLALGHSPFDILMTVRSVLKTLKAP--EHILCEFLKTISLSHMTMISGLS 300
Query 67 TELQME 72
+ LQ+E
Sbjct 301 SPLQLE 306
> pfa:PFL2005w replication factor C subunit 4; K10755 replication
factor C subunit 2/4
Length=336
Score = 52.8 bits (125), Expect = 3e-07, Method: Composition-based stats.
Identities = 30/80 (37%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Query 7 WYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLS 66
W A +A ++ G+TP D+ T VLRR + E + +EFL I M TMA GL+
Sbjct 255 WKKAHDIAYSMIKEGHTPYDISLTSSNVLRRFNIG-SEVIQIEFLKIGAMACNTMATGLT 313
Query 67 TELQMEKMLAQLCKVAISFR 86
+ +Q++K+LA C A R
Sbjct 314 SVIQLDKLLADWCMAAKILR 333
> xla:431883 rfc2, MGC81391, rfc40; replication factor C (activator
1) 2, 40kDa; K10755 replication factor C subunit 2/4
Length=348
Score = 50.4 bits (119), Expect = 1e-06, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 43/66 (65%), Gaps = 3/66 (4%)
Query 20 LGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
LGY+P D++ V + ++ E+L LEF+ +G THM +A G+++ LQM +LA+LC
Sbjct 283 LGYSPEDIIGNIFRVCKTF--QMPEYLKLEFIKEIGYTHMKIAEGVNSLLQMAGLLARLC 340
Query 80 -KVAIS 84
K A+S
Sbjct 341 QKTAVS 346
> hsa:5982 RFC2, A1, MGC3665, RFC40; replication factor C (activator
1) 2, 40kDa; K10755 replication factor C subunit 2/4
Length=320
Score = 49.7 bits (117), Expect = 3e-06, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 40/61 (65%), Gaps = 2/61 (3%)
Query 20 LGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
LGY+P D++ V + ++ E+L LEF+ +G THM +A G+++ LQM +LA+LC
Sbjct 254 LGYSPEDIIGNIFRVCKTF--QMAEYLKLEFIKEIGYTHMKIAEGVNSLLQMAGLLARLC 311
Query 80 K 80
+
Sbjct 312 Q 312
> dre:503748 rfc2, zgc:110810; replication factor C (activator
1) 2; K10755 replication factor C subunit 2/4
Length=349
Score = 48.9 bits (115), Expect = 4e-06, Method: Composition-based stats.
Identities = 24/71 (33%), Positives = 43/71 (60%), Gaps = 2/71 (2%)
Query 10 AQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTEL 69
A + + SLGY+P D++ V + ++ E+L LE++ +G THM +A G+++ L
Sbjct 273 AYKIIEQLWSLGYSPEDIIGNIFRVCKTF--QMAEYLKLEYIKEIGYTHMKVAEGVNSLL 330
Query 70 QMEKMLAQLCK 80
QM +L +LC+
Sbjct 331 QMAGLLGRLCR 341
> mmu:19718 Rfc2, 2610008M13Rik, 40kDa, AI326953, MGC117486, Recc2;
replication factor C (activator 1) 2; K10755 replication
factor C subunit 2/4
Length=349
Score = 48.5 bits (114), Expect = 5e-06, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 39/61 (63%), Gaps = 2/61 (3%)
Query 20 LGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
LGY+P DV+ V + + E+L LEF+ +G THM +A G+++ LQM +LA+LC
Sbjct 283 LGYSPEDVIGNIFRVCKTF--PMAEYLKLEFIKEIGYTHMKVAEGVNSLLQMAGLLARLC 340
Query 80 K 80
+
Sbjct 341 Q 341
> cel:F58F6.4 rfc-2; RFC (DNA replication factor) family member
(rfc-2); K10755 replication factor C subunit 2/4
Length=334
Score = 47.8 bits (112), Expect = 7e-06, Method: Composition-based stats.
Identities = 22/79 (27%), Positives = 46/79 (58%), Gaps = 2/79 (2%)
Query 5 RVWYGAQSVARVILSLGYTPLDVVTTFRGVLRRAD--SELQEHLLLEFLGIVGMTHMTMA 62
R ++ A + LG++ D+V+T V++ + + E L +E++ + M HM +
Sbjct 247 RKFFEASKIIHEFHRLGFSSDDIVSTLFRVVKTVELSKNVSEQLRMEYIRQIAMCHMRIV 306
Query 63 GGLSTELQMEKMLAQLCKV 81
GL+++LQ+ +++A LC+V
Sbjct 307 QGLTSKLQLSRLIADLCRV 325
> ath:AT1G63160 replication factor C 40 kDa, putative; K10755
replication factor C subunit 2/4
Length=333
Score = 47.4 bits (111), Expect = 1e-05, Method: Composition-based stats.
Identities = 23/65 (35%), Positives = 39/65 (60%), Gaps = 2/65 (3%)
Query 17 ILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLA 76
+ LGY+P D++TT +++ D + E+L LEF+ G HM + G+ + LQ+ +LA
Sbjct 264 LYDLGYSPTDIITTLFRIIKNYD--MAEYLKLEFMKETGFAHMRICDGVGSYLQLCGLLA 321
Query 77 QLCKV 81
+L V
Sbjct 322 KLSIV 326
> sce:YOL094C RFC4; Rfc4p; K10755 replication factor C subunit
2/4
Length=323
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 23/62 (37%), Positives = 42/62 (67%), Gaps = 3/62 (4%)
Query 21 GYTPLDVVTT-FRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLC 79
GY+ +D+VTT FR + + ++++E + LE + +G+THM + G+ T LQ+ MLA++
Sbjct 260 GYSSIDIVTTSFR--VTKNLAQVKESVRLEMIKEIGLTHMRILEGVGTYLQLASMLAKIH 317
Query 80 KV 81
K+
Sbjct 318 KL 319
> ath:AT5G53540 MSP1 protein, putative / intramitochondrial sorting
protein, putative
Length=403
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 25/89 (28%), Positives = 39/89 (43%), Gaps = 20/89 (22%)
Query 7 WYG-AQSVARVILSLGYT--P----LDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHM 59
W+G AQ + + SL Y P +D V +F G R D+E ++ EF M
Sbjct 162 WFGDAQKLVSAVFSLAYKLQPAIIFIDEVDSFLGQRRSTDNEAMSNMKTEF--------M 213
Query 60 TMAGGLSTELQMEKMLAQLCKVAISFRPS 88
+ G +T+ M+ +A + RPS
Sbjct 214 ALWDGFTTDQNARVMV-----LAATNRPS 237
Lambda K H
0.326 0.139 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2007827920
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40