bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
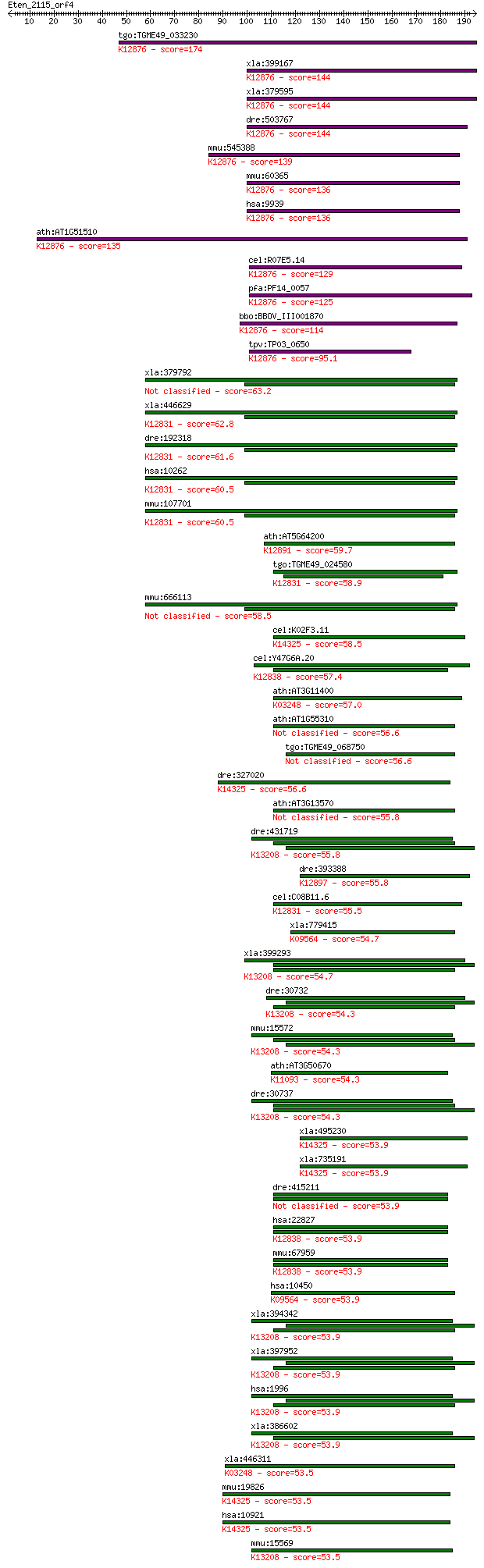
Query= Eten_2115_orf4
Length=194
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_033230 RNA binding motif-containing protein ; K1287... 174 2e-43
xla:399167 rbm8, rbm8-A, rbm8a-A; RNA-binding protein Y14; K12... 144 1e-34
xla:379595 rbm8a-b, MGC68591, rbm8-B; RNA-binding protein 8A-B... 144 1e-34
dre:503767 rbm8a, fb34f03, wu:fb34f03, zgc:110525; RNA binding... 144 2e-34
mmu:545388 B020018G12Rik, Y14; RIKEN cDNA B020018G12 gene; K12... 139 7e-33
mmu:60365 Rbm8a, 2310057C03Rik, AA673428, Rbm8; RNA binding mo... 136 5e-32
hsa:9939 RBM8A, BOV-1A, BOV-1B, BOV-1C, MDS014, RBM8, RBM8B, Y... 136 5e-32
ath:AT1G51510 Y14; Y14; RNA binding / protein binding; K12876 ... 135 1e-31
cel:R07E5.14 rnp-4; RNP (RRM RNA binding domain) containing fa... 129 7e-30
pfa:PF14_0057 RNA binding protein, putative; K12876 RNA-bindin... 125 1e-28
bbo:BBOV_III001870 17.m07184; RNA recognition motif domain con... 114 3e-25
tpv:TP03_0650 hypothetical protein; K12876 RNA-binding protein 8A 95.1 1e-19
xla:379792 sf3b4, MGC52742, spx; splicing factor 3b, subunit 4... 63.2 6e-10
xla:446629 MGC82420 protein; K12831 splicing factor 3B subunit 4 62.8
dre:192318 sf3b4, chunp6902, fb36a04, wu:fb36a04; splicing fac... 61.6 1e-09
hsa:10262 SF3B4, Hsh49, MGC10828, SAP49, SF3b49; splicing fact... 60.5 4e-09
mmu:107701 Sf3b4, 49kDa, MGC36369, MGC36564, SF3b49, SF3b50, S... 60.5 4e-09
ath:AT5G64200 ATSC35; ATSC35; RNA binding / nucleic acid bindi... 59.7 6e-09
tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12... 58.9 1e-08
mmu:666113 Gm7935, EG666113; predicted pseudogene 7935 58.5
cel:K02F3.11 rnp-5; RNP (RRM RNA binding domain) containing fa... 58.5 1e-08
cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing f... 57.4 3e-08
ath:AT3G11400 EIF3G1; eukaryotic translation initiation factor... 57.0 4e-08
ath:AT1G55310 SR33; SR33; RNA binding / protein binding 56.6 6e-08
tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative 56.6 6e-08
dre:327020 rnps1, wu:fa19a11, zgc:65775; RNA binding protein S... 56.6 6e-08
ath:AT3G13570 SCL30a; SCL30a; RNA binding / nucleic acid bindi... 55.8 8e-08
dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal... 55.8 9e-08
dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha; K1... 55.8 1e-07
cel:C08B11.6 actin; Spliceosome-Associated Protein family memb... 55.5 1e-07
xla:779415 ppie, MGC154898, cyp-33, cyp33, cype; peptidylproly... 54.7 2e-07
xla:399293 elavl3, elrC; ELAV (embryonic lethal, abnormal visi... 54.7 2e-07
dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELA... 54.3 3e-07
mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abn... 54.3 3e-07
ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTE... 54.3 3e-07
dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic leth... 54.3 3e-07
xla:495230 rnps1-a; RNA binding protein S1, serine-rich domain... 53.9 3e-07
xla:735191 rnps1-b, MGC131270; RNA binding protein S1, serine-... 53.9 3e-07
dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.... 53.9 3e-07
hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding... 53.9 3e-07
mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U... 53.9 3e-07
hsa:10450 PPIE, CYP-33, CYP33, MGC111222, MGC3736; peptidylpro... 53.9 3e-07
xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,... 53.9 3e-07
xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1; ... 53.9 3e-07
hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal v... 53.9 4e-07
xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic le... 53.9 4e-07
xla:446311 eif3s4, MGC83369, eIF3g-A, eif3s4-A; eukaryotic tra... 53.5 4e-07
mmu:19826 Rnps1; ribonucleic acid binding protein S1; K14325 R... 53.5 4e-07
hsa:10921 RNPS1, E5.1, MGC117332; RNA binding protein S1, seri... 53.5 4e-07
mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnorma... 53.5 4e-07
> tgo:TGME49_033230 RNA binding motif-containing protein ; K12876
RNA-binding protein 8A
Length=162
Score = 174 bits (441), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 99/153 (64%), Positives = 120/153 (78%), Gaps = 5/153 (3%)
Query 47 DRRSASLSRGLRQKGRGMREGVSSRDDRYAGQGGVFERLEE----TAEGKRRGKDKGPAP 102
DRRS S S+ +QKGRG++EG D RY+GQ GVFERLEE T+ + K KG AP
Sbjct 9 DRRSPSASKSRKQKGRGLKEGGRETDGRYSGQAGVFERLEEQNGATSTRRESEKSKGAAP 68
Query 103 ARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDE 162
ARSVEGWI+V N+HEEAQEEDIHE F++FG IKNLH+NLDRR+GFVKGYAF+E+ET++E
Sbjct 69 ARSVEGWIIVVRNLHEEAQEEDIHENFESFGQIKNLHLNLDRRTGFVKGYAFIEFETFEE 128
Query 163 AQAAIEGMDGQELLGQKVAVSWAFVRQP-EKRK 194
A+AAI GMD Q+LLGQ V V WAF + P EKR+
Sbjct 129 AEAAIRGMDNQQLLGQTVYVDWAFSKAPNEKRR 161
> xla:399167 rbm8, rbm8-A, rbm8a-A; RNA-binding protein Y14; K12876
RNA-binding protein 8A
Length=174
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 63/95 (66%), Positives = 76/95 (80%), Gaps = 0/95 (0%)
Query 100 PAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYET 159
P P RSVEGWI+ T +HEEA EEDIH+ F FG IKN+H+NLDRR+GF+KGYA VEYET
Sbjct 64 PGPQRSVEGWILFVTGVHEEATEEDIHDKFGEFGEIKNIHLNLDRRTGFLKGYALVEYET 123
Query 160 YDEAQAAIEGMDGQELLGQKVAVSWAFVRQPEKRK 194
Y EA AA+EG++GQ+L+GQ V+V W FVR P K K
Sbjct 124 YKEALAAMEGLNGQDLMGQPVSVDWGFVRGPPKGK 158
> xla:379595 rbm8a-b, MGC68591, rbm8-B; RNA-binding protein 8A-B;
K12876 RNA-binding protein 8A
Length=174
Score = 144 bits (364), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 63/95 (66%), Positives = 76/95 (80%), Gaps = 0/95 (0%)
Query 100 PAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYET 159
P P RSVEGWI+ T +HEEA EEDIH+ F FG IKN+H+NLDRR+GF+KGYA VEYET
Sbjct 64 PGPQRSVEGWILFVTGVHEEATEEDIHDKFGEFGEIKNIHLNLDRRTGFLKGYALVEYET 123
Query 160 YDEAQAAIEGMDGQELLGQKVAVSWAFVRQPEKRK 194
Y EA AA+EG++GQ+L+GQ V+V W FVR P K K
Sbjct 124 YKEALAAMEGLNGQDLMGQPVSVDWGFVRGPPKGK 158
> dre:503767 rbm8a, fb34f03, wu:fb34f03, zgc:110525; RNA binding
motif protein 8A; K12876 RNA-binding protein 8A
Length=174
Score = 144 bits (363), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 61/91 (67%), Positives = 75/91 (82%), Gaps = 0/91 (0%)
Query 100 PAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYET 159
P P RSVEGWI+ T +HEEA EED+H+ F FG IKNLH+NLDRR+G++KGYA VEYET
Sbjct 64 PGPQRSVEGWILFVTGVHEEATEEDVHDKFAEFGEIKNLHLNLDRRTGYLKGYALVEYET 123
Query 160 YDEAQAAIEGMDGQELLGQKVAVSWAFVRQP 190
Y EAQAA+EG++GQEL+GQ ++V W FVR P
Sbjct 124 YKEAQAAMEGLNGQELMGQPISVDWCFVRGP 154
> mmu:545388 B020018G12Rik, Y14; RIKEN cDNA B020018G12 gene; K12876
RNA-binding protein 8A
Length=174
Score = 139 bits (349), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 59/104 (56%), Positives = 81/104 (77%), Gaps = 1/104 (0%)
Query 84 RLEETAEGKRRGKDKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLD 143
R++E + ++ D+ P P RSVEGWI+ T +HEEA EEDIH+ F +G IKN+H+NLD
Sbjct 49 RMQEDYDSVKQDSDE-PGPQRSVEGWILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLD 107
Query 144 RRSGFVKGYAFVEYETYDEAQAAIEGMDGQELLGQKVAVSWAFV 187
RR+G++KGY VEYETY EAQAA+EG++GQ+L+GQ ++V W FV
Sbjct 108 RRTGYLKGYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFV 151
> mmu:60365 Rbm8a, 2310057C03Rik, AA673428, Rbm8; RNA binding
motif protein 8a; K12876 RNA-binding protein 8A
Length=174
Score = 136 bits (342), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 72/88 (81%), Gaps = 0/88 (0%)
Query 100 PAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYET 159
P P RSVEGWI+ T +HEEA EEDIH+ F +G IKN+H+NLDRR+G++KGY VEYET
Sbjct 64 PGPQRSVEGWILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLDRRTGYLKGYTLVEYET 123
Query 160 YDEAQAAIEGMDGQELLGQKVAVSWAFV 187
Y EAQAA+EG++GQ+L+GQ ++V W FV
Sbjct 124 YKEAQAAMEGLNGQDLMGQPISVDWCFV 151
> hsa:9939 RBM8A, BOV-1A, BOV-1B, BOV-1C, MDS014, RBM8, RBM8B,
Y14, ZNRP, ZRNP1; RNA binding motif protein 8A; K12876 RNA-binding
protein 8A
Length=174
Score = 136 bits (342), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 56/88 (63%), Positives = 72/88 (81%), Gaps = 0/88 (0%)
Query 100 PAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYET 159
P P RSVEGWI+ T +HEEA EEDIH+ F +G IKN+H+NLDRR+G++KGY VEYET
Sbjct 64 PGPQRSVEGWILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLDRRTGYLKGYTLVEYET 123
Query 160 YDEAQAAIEGMDGQELLGQKVAVSWAFV 187
Y EAQAA+EG++GQ+L+GQ ++V W FV
Sbjct 124 YKEAQAAMEGLNGQDLMGQPISVDWCFV 151
> ath:AT1G51510 Y14; Y14; RNA binding / protein binding; K12876
RNA-binding protein 8A
Length=202
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 82/186 (44%), Positives = 100/186 (53%), Gaps = 39/186 (20%)
Query 13 QGRIAAGAAVPAAAAAAARRTAAAAAAAEMAEAEDRRSASLSRGLRQKGRGMREG----- 67
+G GA V A ++A A A E A+ + KGRG RE
Sbjct 22 EGTAIDGADVSPRAGHPRLKSAIAGANGESAK-------------KTKGRGFREEKDSDR 68
Query 68 ---VSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVEGWIVVATNIHEEAQEED 124
+SSRD FE L G D P P RSVEGWI++ + +HEE QEED
Sbjct 69 QRRLSSRD---------FESL---------GSDGRPGPQRSVEGWIILVSGVHEETQEED 110
Query 125 IHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQELLGQKVAVSW 184
I AF FG IKNL++NLDRRSG+VKGYA +EYE +EAQ+AI M+G ELL Q V+V W
Sbjct 111 ITNAFGDFGEIKNLNLNLDRRSGYVKGYALIEYEKKEEAQSAISAMNGAELLTQNVSVDW 170
Query 185 AFVRQP 190
AF P
Sbjct 171 AFSSGP 176
> cel:R07E5.14 rnp-4; RNP (RRM RNA binding domain) containing
family member (rnp-4); K12876 RNA-binding protein 8A
Length=142
Score = 129 bits (324), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 55/88 (62%), Positives = 67/88 (76%), Gaps = 0/88 (0%)
Query 101 APARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETY 160
AP RSVEGWIV TNIHEEA E+D+H+ F +G IKN+H+NLDRR+GF+KGYA VEYET
Sbjct 47 APQRSVEGWIVFVTNIHEEATEDDVHDKFSEYGKIKNIHLNLDRRTGFLKGYALVEYETQ 106
Query 161 DEAQAAIEGMDGQELLGQKVAVSWAFVR 188
EA AI+ + +LLGQ V V W FV+
Sbjct 107 KEANEAIDQSNDTDLLGQNVKVDWCFVK 134
> pfa:PF14_0057 RNA binding protein, putative; K12876 RNA-binding
protein 8A
Length=107
Score = 125 bits (313), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 53/92 (57%), Positives = 73/92 (79%), Gaps = 0/92 (0%)
Query 101 APARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETY 160
PA+SVEGWI++ TNIH EA+++ I E F+ FG IKNLH+N+DRR+GF+KGYAF+EYE +
Sbjct 15 VPAKSVEGWIIIITNIHGEARDDYIKEVFERFGQIKNLHLNIDRRTGFLKGYAFLEYENF 74
Query 161 DEAQAAIEGMDGQELLGQKVAVSWAFVRQPEK 192
+A+ AI+ MDG LL Q++ V WAFV++ K
Sbjct 75 VDAKRAIDEMDGTMLLNQEIHVDWAFVQEKSK 106
> bbo:BBOV_III001870 17.m07184; RNA recognition motif domain containing
protein; K12876 RNA-binding protein 8A
Length=98
Score = 114 bits (284), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 46/90 (51%), Positives = 69/90 (76%), Gaps = 0/90 (0%)
Query 97 DKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVE 156
D P +++ GW+++ T +H EAQEEDI +AF+ FG I ++H+N+DRRSG+ KGYAF+E
Sbjct 2 DNEEVPIKTIGGWVIIVTGVHPEAQEEDIRDAFEGFGQITSIHLNMDRRSGYAKGYAFIE 61
Query 157 YETYDEAQAAIEGMDGQELLGQKVAVSWAF 186
+ EA++AI M+G ++LGQ+VAV+WAF
Sbjct 62 FSEQQEARSAITEMNGHQILGQEVAVAWAF 91
> tpv:TP03_0650 hypothetical protein; K12876 RNA-binding protein
8A
Length=76
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 37/67 (55%), Positives = 56/67 (83%), Gaps = 0/67 (0%)
Query 101 APARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETY 160
AP +S+EGW+++ T IHEEAQ+ED+ EAF+ +G + NLH+NLDRR+G+VKGYA +E++
Sbjct 7 APIKSIEGWVLMVTGIHEEAQDEDVREAFENYGTVTNLHLNLDRRTGYVKGYALLEFKNK 66
Query 161 DEAQAAI 167
+EA+ A+
Sbjct 67 EEAENAM 73
> xla:379792 sf3b4, MGC52742, spx; splicing factor 3b, subunit
4, 49kDa
Length=377
Score = 63.2 bits (152), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGYAF+ Y ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINYASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> xla:446629 MGC82420 protein; K12831 splicing factor 3B subunit
4
Length=383
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGYAF+ Y ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINYASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> dre:192318 sf3b4, chunp6902, fb36a04, wu:fb36a04; splicing factor
3b, subunit 4; K12831 splicing factor 3B subunit 4
Length=400
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGYAF+ + ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINFASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> hsa:10262 SF3B4, Hsh49, MGC10828, SAP49, SF3b49; splicing factor
3b, subunit 4, 49kDa; K12831 splicing factor 3B subunit
4
Length=424
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGYAF+ + ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINFASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> mmu:107701 Sf3b4, 49kDa, MGC36369, MGC36564, SF3b49, SF3b50,
Sap49; splicing factor 3b, subunit 4; K12831 splicing factor
3B subunit 4
Length=424
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGYAF+ + ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFSAFGVILQTPKIMRDPDTGNSKGYAFINFASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> ath:AT5G64200 ATSC35; ATSC35; RNA binding / nucleic acid binding
/ nucleotide binding; K12891 splicing factor, arginine/serine-rich
2
Length=303
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 48/79 (60%), Gaps = 0/79 (0%)
Query 107 EGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAA 166
+ + ++ NI +D++ F +G + ++ + DRR+G +G+AFV Y+ DEA A
Sbjct 14 DTYSLLVLNITFRTTADDLYPLFAKYGKVVDVFIPRDRRTGDSRGFAFVRYKYKDEAHKA 73
Query 167 IEGMDGQELLGQKVAVSWA 185
+E +DG+ + G+++ V +A
Sbjct 74 VERLDGRVVDGREITVQFA 92
> tgo:TGME49_024580 splicing factor, putative (EC:3.1.2.15); K12831
splicing factor 3B subunit 4
Length=576
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 45/76 (59%), Gaps = 0/76 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V N+ + E+ I++ F AFG I + + D +G +G+ FV ++T++ + AA+ M
Sbjct 118 VFLGNLDPDVDEKTIYDTFSAFGNIISAKIMRDPETGLSRGFGFVSFDTFEASDAALAAM 177
Query 171 DGQELLGQKVAVSWAF 186
+GQ + + + VS+A+
Sbjct 178 NGQFICNRPIHVSYAY 193
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 36/66 (54%), Gaps = 0/66 (0%)
Query 115 NIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQE 174
N+ + ++ + E F G ++ + + D+ +G +GY FVE+ +A A++ M+ +
Sbjct 35 NLDSQVDDDLLWELFVQCGPVRTVSVPRDKLTGNHQGYGFVEFRNEVDADYALKLMNMVK 94
Query 175 LLGQKV 180
L G+ +
Sbjct 95 LYGKAL 100
> mmu:666113 Gm7935, EG666113; predicted pseudogene 7935
Length=425
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 40/131 (30%), Positives = 69/131 (52%), Gaps = 6/131 (4%)
Query 58 RQKGRGMREGVSSRDDRYAGQGGVFERLEETAEGKRRGKDKGPAPARSVE-GWIVVATNI 116
+ +G G E +S D YA + +L GK +K A ++++ G + N+
Sbjct 52 QHQGYGFVEFLSEEDADYAIKIMNMIKLY----GKPIRVNKASAHNKNLDVGANIFIGNL 107
Query 117 HEEAQEEDIHEAFDAFGAI-KNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
E E+ +++ F AFG I + + D +G KGY+F+ + ++D + AAIE M+GQ L
Sbjct 108 DPEIDEKLLYDTFRAFGVILQTPKIMWDPDTGNSKGYSFINFASFDASDAAIEAMNGQYL 167
Query 176 LGQKVAVSWAF 186
+ + VS+AF
Sbjct 168 CNRPITVSYAF 178
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 47/87 (54%), Gaps = 1/87 (1%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP R+ + + V + E+ E + E F G + N HM DR +G +GY FVE+
Sbjct 4 GPISERNQDATVYVG-GLDEKVSEPLLWELFLQAGPVVNTHMPKDRVTGQHQGYGFVEFL 62
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWA 185
+ ++A AI+ M+ +L G+ + V+ A
Sbjct 63 SEEDADYAIKIMNMIKLYGKPIRVNKA 89
> cel:K02F3.11 rnp-5; RNP (RRM RNA binding domain) containing
family member (rnp-5); K14325 RNA-binding protein with serine-rich
domain 1
Length=289
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 28/80 (35%), Positives = 44/80 (55%), Gaps = 1/80 (1%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFV-KGYAFVEYETYDEAQAAIEG 169
VV N+ + + E F +GAIKN+ + DR V +GY +VEY+ ++A+ +I+
Sbjct 135 VVIKNLSRNVLKTHLEEIFSIYGAIKNVDLPPDRFHNHVHRGYGYVEYDNLEDAEKSIKH 194
Query 170 MDGQELLGQKVAVSWAFVRQ 189
MDG ++ G V V RQ
Sbjct 195 MDGGQIDGMAVHVEMTIGRQ 214
> cel:Y47G6A.20 rnp-6; RNP (RRM RNA binding domain) containing
family member (rnp-6); K12838 poly(U)-binding-splicing factor
PUF60
Length=749
Score = 57.4 bits (137), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 37/92 (40%), Positives = 54/92 (58%), Gaps = 4/92 (4%)
Query 103 ARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDE 162
A S+ I V + I E +E+ + AFD FG IK+++M+ D +G K +AFVEYE +
Sbjct 97 ALSIMSRIYVGS-ISFEIREDMLRRAFDPFGPIKSINMSWDPATGHHKTFAFVEYEVPEA 155
Query 163 AQAAIEGMDGQELLGQKVAV-SWAF--VRQPE 191
A A E M+GQ L G+ + V S F +R P+
Sbjct 156 ALLAQESMNGQMLGGRNLKVNSMMFQEMRLPQ 187
Score = 35.8 bits (81), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 19/72 (26%), Positives = 34/72 (47%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V +++H + E D+ F+AFG I + +G+ ++E+ AI GM
Sbjct 209 VYVSSVHPDLSETDLKSVFEAFGEIVKCQLARAPTGRGHRGFGYLEFNNLTSQSEAIAGM 268
Query 171 DGQELLGQKVAV 182
+ +L GQ + V
Sbjct 269 NMFDLGGQYLRV 280
> ath:AT3G11400 EIF3G1; eukaryotic translation initiation factor
3G / eIF3g; K03248 translation initiation factor 3 subunit
G
Length=321
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 25/78 (32%), Positives = 46/78 (58%), Gaps = 0/78 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V TN+ E+ +E D+ E F FGA+ +++ +D+++G +G+ FV + + ++AQ AI +
Sbjct 242 VRVTNLSEDTREPDLMELFHPFGAVTRVYVAIDQKTGVSRGFGFVNFVSREDAQRAINKL 301
Query 171 DGQELLGQKVAVSWAFVR 188
+G + V WA R
Sbjct 302 NGYGYDNLILRVEWATPR 319
> ath:AT1G55310 SR33; SR33; RNA binding / protein binding
Length=220
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 47/75 (62%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ N+ + ++ED+ ++F+ FG +K++++ D +G +G+ FV++ +A A M
Sbjct 38 LLVRNLRHDCRQEDLRKSFEQFGPVKDIYLPRDYYTGDPRGFGFVQFMDPADAADAKHHM 97
Query 171 DGQELLGQKVAVSWA 185
DG LLG+++ V +A
Sbjct 98 DGYLLLGRELTVVFA 112
> tgo:TGME49_068750 peptidyl-prolyl cis-trans isomerase E, putative
Length=145
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 44/70 (62%), Gaps = 0/70 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ E+ +EE + AF FG IK L + D+ +G +G+ FVE+E D+A+ A+E MD EL
Sbjct 28 LAEQVEEEVLRAAFLPFGDIKQLEIPKDKTTGLHRGFGFVEFEEEDDAKEAMENMDNAEL 87
Query 176 LGQKVAVSWA 185
G+ + V+ +
Sbjct 88 YGRTLRVNLS 97
> dre:327020 rnps1, wu:fa19a11, zgc:65775; RNA binding protein
S1, serine-rich domain; K14325 RNA-binding protein with serine-rich
domain 1
Length=283
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 32/97 (32%), Positives = 52/97 (53%), Gaps = 7/97 (7%)
Query 88 TAEGKRRGKDKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSG 147
T E +R+ + P P + G + ++ I E F +G IK + M DR
Sbjct 122 TDEKERKRRSPSPKPTKLYLGRLT------RNVTKDHIQEIFATYGKIKMIDMPSDRLHP 175
Query 148 FV-KGYAFVEYETYDEAQAAIEGMDGQELLGQKVAVS 183
V KGYA+VEYE+ ++AQ A++ MDG ++ GQ++ +
Sbjct 176 NVSKGYAYVEYESPEDAQKALKHMDGGQIDGQEITAT 212
> ath:AT3G13570 SCL30a; SCL30a; RNA binding / nucleic acid binding
/ nucleotide binding
Length=262
Score = 55.8 bits (133), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 45/75 (60%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ N+ + ++ED+ F+ FG +K++++ D +G +G+ F+++ +A A M
Sbjct 39 LLVRNLRHDCRQEDLRRPFEQFGPVKDIYLPRDYYTGDPRGFGFIQFMDPADAAEAKHQM 98
Query 171 DGQELLGQKVAVSWA 185
DG LLG+++ V +A
Sbjct 99 DGYLLLGRELTVVFA 113
> dre:431719 elavl2, zgc:91918; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=360
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 43/83 (51%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
PA + GW + N+ +A E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 270 PAHAGTGWCIFVYNLAPDADENVLWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYD 329
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 330 EAAVAIASLNGYRLGDRVLQVSF 352
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++++ + F FG I + +D+ +G +G F+ ++ EA+ AI+G++GQ+
Sbjct 132 LPKTMTQKELEQLFSQFGRIITSRILVDQVTGVSRGVGFIRFDRRVEAEEAIKGLNGQKP 191
Query 176 LGQKVAVSWAFVRQPEKR 193
G ++ F P ++
Sbjct 192 PGATEPITVKFANNPSQK 209
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E++ F + G I++ + D+ +G GY FV Y +A+ AI +
Sbjct 41 LIVNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYMEPKDAEKAINTL 100
Query 171 DGQELLGQKVAVSWA 185
+G L + + VS+A
Sbjct 101 NGLRLQTKTIKVSYA 115
> dre:393388 tra2a, MGC64175, zgc:64175; transformer-2 alpha;
K12897 transformer-2 protein
Length=297
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 46/70 (65%), Gaps = 0/70 (0%)
Query 122 EEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQELLGQKVA 181
E D+ E F +G++ +++ D+R+G +G+AFV +E D+A+ A+E +G EL G+++
Sbjct 148 ERDLREVFSRYGSLAGVNVVYDQRTGRSRGFAFVYFEHIDDAKEAMERANGMELDGRRIR 207
Query 182 VSWAFVRQPE 191
V ++ ++P
Sbjct 208 VDYSITKRPH 217
> cel:C08B11.6 actin; Spliceosome-Associated Protein family member
(sap-49); K12831 splicing factor 3B subunit 4
Length=388
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 26/79 (32%), Positives = 46/79 (58%), Gaps = 1/79 (1%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNL-DRRSGFVKGYAFVEYETYDEAQAAIEG 169
+ N+ E E+ +++ F AFG I + + D SG KG+AF+ + +++ + A+E
Sbjct 102 IFVGNLDPEVDEKLLYDTFSAFGVILQVPKIMRDVDSGTSKGFAFINFASFEASDTALEA 161
Query 170 MDGQELLGQKVAVSWAFVR 188
M+GQ L + + VS+AF R
Sbjct 162 MNGQFLCNRAITVSYAFKR 180
> xla:779415 ppie, MGC154898, cyp-33, cyp33, cype; peptidylprolyl
isomerase E (cyclophilin E); K09564 peptidyl-prolyl isomerase
E (cyclophilin E) [EC:5.2.1.8]
Length=294
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/68 (39%), Positives = 43/68 (63%), Gaps = 0/68 (0%)
Query 118 EEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQELLG 177
EE E+ +H AF FG I ++ + LD + +G+AFVE+E ++A AAI+ M+ EL G
Sbjct 8 EEVDEKILHAAFIPFGDITDIQIPLDYETEKHRGFAFVEFELPEDAAAAIDNMNESELFG 67
Query 178 QKVAVSWA 185
+ + V+ A
Sbjct 68 RTIRVNLA 75
> xla:399293 elavl3, elrC; ELAV (embryonic lethal, abnormal vision)-like
3 (Hu antigen C); K13208 ELAV like protein 2/3/4
Length=348
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 30/91 (32%), Positives = 45/91 (49%), Gaps = 4/91 (4%)
Query 99 GPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYE 158
GP A GW + N+ EA E + + F FGA+ N+ + D + KG+ FV
Sbjct 259 GPTTA----GWCIFVYNLSPEADESVLWQLFGPFGAVTNVKVIRDFTTNKCKGFGFVTMT 314
Query 159 TYDEAQAAIEGMDGQELLGQKVAVSWAFVRQ 189
YDEA AI ++G L + + VS+ +Q
Sbjct 315 NYDEAAMAIASLNGYRLGDRVLQVSFKTSKQ 345
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 18/83 (21%), Positives = 46/83 (55%), Gaps = 0/83 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
+ +++ + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G+
Sbjct 122 LYVSSLPKTMNQKEMEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGL 181
Query 171 DGQELLGQKVAVSWAFVRQPEKR 193
+GQ+ LG ++ F P ++
Sbjct 182 NGQKPLGASEPITVKFANNPSQK 204
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E+ F + G I++ + D+ +G GY FV Y ++A AI +
Sbjct 36 LIVNYLPQNMTQEEFKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYVDPNDADKAINTL 95
Query 171 DGQELLGQKVAVSWA 185
+G +L + + VS+A
Sbjct 96 NGLKLQTKTIKVSYA 110
> dre:30732 elavl3, HuC, elrc, id:ibd1248, wu:fb77b03, zHuC; ELAV
(embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu
antigen C); K13208 ELAV like protein 2/3/4
Length=345
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 42/82 (51%), Gaps = 0/82 (0%)
Query 108 GWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAI 167
GW + N+ EA E + + F FGA+ N+ + D + KG+ FV YDEA AI
Sbjct 261 GWCIFVYNLSPEADESVLWQLFGPFGAVTNVKVIRDFTTNKCKGFGFVTMTNYDEAAMAI 320
Query 168 EGMDGQELLGQKVAVSWAFVRQ 189
++G L + + VS+ +Q
Sbjct 321 ASLNGYRLGDRVLQVSFKTSKQ 342
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 44/78 (56%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++D+ + F +G I + +++ +G +G F+ ++ +EA+ AI+G++GQ+
Sbjct 131 LPKTMSQKDMEQLFSQYGRIITSRILVNQVTGISRGVGFIRFDKRNEAEEAIKGLNGQKP 190
Query 176 LGQKVAVSWAFVRQPEKR 193
LG ++ F P ++
Sbjct 191 LGAAEPITVKFANNPSQK 208
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E+ F + G I++ + D+ +G GY FV Y ++A AI +
Sbjct 40 LIVNYLPQNMTQEEFKSLFGSIGEIESCKLVRDKITGQSLGYGFVNYVDPNDADKAINTL 99
Query 171 DGQELLGQKVAVSWA 185
+G +L + + VS+A
Sbjct 100 NGLKLQTKTIKVSYA 114
> mmu:15572 Elavl4, Elav, Hud, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=380
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ ++ E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 290 PGHTGTGWCIFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYD 349
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 350 EAAMAIASLNGYRLGDRVLQVSF 372
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/78 (21%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G++GQ+
Sbjct 139 LPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGLNGQKP 198
Query 176 LGQKVAVSWAFVRQPEKR 193
G ++ F P ++
Sbjct 199 SGATEPITVKFANNPSQK 216
Score = 40.0 bits (92), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E+ F + G I++ + D+ +G GY FV Y +A+ AI +
Sbjct 48 LIVNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTL 107
Query 171 DGQELLGQKVAVSWA 185
+G L + + VS+A
Sbjct 108 NGLRLQTKTIKVSYA 122
> ath:AT3G50670 U1-70K; U1-70K (U1 SMALL NUCLEAR RIBONUCLEOPROTEIN-70K);
RNA binding / nucleic acid binding / nucleotide binding;
K11093 U1 small nuclear ribonucleoprotein 70kDa
Length=427
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 44/73 (60%), Gaps = 0/73 (0%)
Query 110 IVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEG 169
+ + ++ E+ E I F+++G IK +H+ D+ + KGYAF+EY + +AA +
Sbjct 139 TLFVSRLNYESSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIEYMHTRDMKAAYKQ 198
Query 170 MDGQELLGQKVAV 182
DGQ++ G++V V
Sbjct 199 ADGQKIDGRRVLV 211
> dre:30737 elavl4, elrd, wu:fc24h01, zHuD; ELAV (embryonic lethal,
abnormal vision, Drosophila)-like 4 (Hu antigen D); K13208
ELAV like protein 2/3/4
Length=367
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ ++ E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 277 PGHTGTGWCIFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYD 336
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 337 EAAMAIASLNGYRLGDRVLQVSF 359
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E+ F + G I++ + D+ +G GY FV Y +A+ AI +
Sbjct 46 LIVNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTL 105
Query 171 DGQELLGQKVAVSWA 185
+G L + + VS+A
Sbjct 106 NGLRLQTKTIKVSYA 120
Score = 33.5 bits (75), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 17/86 (19%), Positives = 44/86 (51%), Gaps = 3/86 (3%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFV---KGYAFVEYETYDEAQAAI 167
+ + + + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI
Sbjct 132 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGPTGGSRGVGFIRFDKRIEAEEAI 191
Query 168 EGMDGQELLGQKVAVSWAFVRQPEKR 193
+G++GQ+ G ++ F P ++
Sbjct 192 KGLNGQKPSGAAEPITVKFANNPSQK 217
> xla:495230 rnps1-a; RNA binding protein S1, serine-rich domain;
K14325 RNA-binding protein with serine-rich domain 1
Length=283
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 41/70 (58%), Gaps = 1/70 (1%)
Query 122 EEDIHEAFDAFGAIKNLHMNLDR-RSGFVKGYAFVEYETYDEAQAAIEGMDGQELLGQKV 180
++ I E F +G IK + M +DR KGYA+VE+E +EA+ A++ MDG ++ GQ++
Sbjct 151 KDHILEIFSTYGKIKMIDMPVDRYHPHLSKGYAYVEFEAPEEAEKALKHMDGGQIDGQEI 210
Query 181 AVSWAFVRQP 190
S P
Sbjct 211 TASAVLTPWP 220
> xla:735191 rnps1-b, MGC131270; RNA binding protein S1, serine-rich
domain; K14325 RNA-binding protein with serine-rich domain
1
Length=283
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 41/70 (58%), Gaps = 1/70 (1%)
Query 122 EEDIHEAFDAFGAIKNLHMNLDR-RSGFVKGYAFVEYETYDEAQAAIEGMDGQELLGQKV 180
++ I E F +G IK + M +DR KGYA+VE+E +EA+ A++ MDG ++ GQ++
Sbjct 151 KDHILEIFSTYGKIKMIDMPVDRYHPHLSKGYAYVEFEAPEEAEKALKHMDGGQIDGQEI 210
Query 181 AVSWAFVRQP 190
S P
Sbjct 211 TASAVLTPWP 220
> dre:415211 puf60b, SIAHBP1, fe37c05, repressor, si:ch211-12p8.2,
si:zc12p8.2, wu:fb33e11, wu:fe37c05, zgc:86806; poly-U
binding splicing factor b
Length=502
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V +I+ E E+ I +AF FG IK++ M+ D + KG+AFVEYE + AQ A+E M
Sbjct 71 VYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQM 130
Query 171 DGQELLGQKVAV 182
+ L G+ + V
Sbjct 131 NSVMLGGRNIKV 142
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 40/72 (55%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
+ +IH + ++DI F+AFG IK+ + + +G KG+ F+EYE + A+ M
Sbjct 168 IYVASIHPDLSDDDIKSVFEAFGKIKSCMLAREPTTGKHKGFGFIEYEKPQSSLDAVSSM 227
Query 171 DGQELLGQKVAV 182
+ +L GQ + V
Sbjct 228 NLFDLGGQYLRV 239
> hsa:22827 PUF60, FIR, FLJ31379, RoBPI, SIAHBP1; poly-U binding
splicing factor 60KDa; K12838 poly(U)-binding-splicing factor
PUF60
Length=516
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V +I+ E E+ I +AF FG IK++ M+ D + KG+AFVEYE + AQ A+E M
Sbjct 88 VYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQM 147
Query 171 DGQELLGQKVAV 182
+ L G+ + V
Sbjct 148 NSVMLGGRNIKV 159
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
+ ++H++ ++DI F+AFG IK+ + D +G KGY F+EYE +Q A+ M
Sbjct 185 IYVASVHQDLSDDDIKSVFEAFGKIKSCTLARDPTTGKHKGYGFIEYEKAQSSQDAVSSM 244
Query 171 DGQELLGQKVAV 182
+ +L GQ + V
Sbjct 245 NLFDLGGQYLRV 256
> mmu:67959 Puf60, 2410104I19Rik, 2810454F19Rik, SIAHBP1; poly-U
binding splicing factor 60; K12838 poly(U)-binding-splicing
factor PUF60
Length=499
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
V +I+ E E+ I +AF FG IK++ M+ D + KG+AFVEYE + AQ A+E M
Sbjct 71 VYVGSIYYELGEDTIRQAFAPFGPIKSIDMSWDSVTMKHKGFAFVEYEVPEAAQLALEQM 130
Query 171 DGQELLGQKVAV 182
+ L G+ + V
Sbjct 131 NSVMLGGRNIKV 142
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
+ ++H++ ++DI F+AFG IK+ + D +G KGY F+EYE +Q A+ M
Sbjct 168 IYVASVHQDLSDDDIKSVFEAFGKIKSCTLARDPTTGKHKGYGFIEYEKAQSSQDAVSSM 227
Query 171 DGQELLGQKVAV 182
+ +L GQ + V
Sbjct 228 NLFDLGGQYLRV 239
> hsa:10450 PPIE, CYP-33, CYP33, MGC111222, MGC3736; peptidylprolyl
isomerase E (cyclophilin E) (EC:5.2.1.8); K09564 peptidyl-prolyl
isomerase E (cyclophilin E) [EC:5.2.1.8]
Length=314
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/76 (34%), Positives = 46/76 (60%), Gaps = 0/76 (0%)
Query 110 IVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEG 169
++ + EE ++ +H AF FG I ++ + LD + +G+AFVE+E ++A AAI+
Sbjct 7 VLYVGGLAEEVDDKVLHAAFIPFGDITDIQIPLDYETEKHRGFAFVEFELAEDAAAAIDN 66
Query 170 MDGQELLGQKVAVSWA 185
M+ EL G+ + V+ A
Sbjct 67 MNESELFGRTIRVNLA 82
> xla:394342 elavl2-a, Xel-1, el-1, elavl2, elrB, hel-n1, heln1,
hub, p45; ELAV (embryonic lethal, abnormal vision)-like 2
(Hu antigen B); K13208 ELAV like protein 2/3/4
Length=389
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ +A E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 299 PGHAGTGWCIFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYD 358
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 359 EAAMAIASLNGYRLGDRVLQVSF 381
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/78 (21%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G++GQ+
Sbjct 160 LPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGLNGQKP 219
Query 176 LGQKVAVSWAFVRQPEKR 193
G ++ F P ++
Sbjct 220 PGATEPITVKFANNPSQK 237
Score = 35.8 bits (81), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRS-GFVKGYAFVEYETYDEAQAAIEG 169
++ + + +E++ F + G I++ + D+ + G GY FV Y +A+ AI
Sbjct 68 LIVNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITEGQSLGYGFVNYIDPKDAEKAINT 127
Query 170 MDGQELLGQKVAVSWA 185
++G L + + VS+A
Sbjct 128 LNGLRLQTKTIKVSYA 143
> xla:397952 elavl2-b, elavl2, elrB, hel-n1, heln1, hub, xel-1;
ELAV (embryonic lethal, abnormal vision)-like 2 (Hu antigen
B); K13208 ELAV like protein 2/3/4
Length=389
Score = 53.9 bits (128), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ +A E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 299 PGHAGTGWCIFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYD 358
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 359 EAAMAIASLNGYRLGDRVLQVSF 381
Score = 39.7 bits (91), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 17/78 (21%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G++GQ+
Sbjct 160 LPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGLNGQKP 219
Query 176 LGQKVAVSWAFVRQPEKR 193
G ++ F P ++
Sbjct 220 PGATEPITVKFANNPSQK 237
Score = 35.0 bits (79), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRS-GFVKGYAFVEYETYDEAQAAIEG 169
++ + + +E++ F + G I++ + D+ + G GY FV Y +A+ AI
Sbjct 68 LIVNYLPQNMTQEELKSLFGSIGEIESCKLVRDKITEGQSLGYGFVNYIDPKDAEKAINT 127
Query 170 MDGQELLGQKVAVSWA 185
++G L + + VS+A
Sbjct 128 VNGLRLQTKTIKVSYA 143
> hsa:1996 ELAVL4, HUD, PNEM; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 4 (Hu antigen D); K13208 ELAV like
protein 2/3/4
Length=366
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ ++ E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 276 PGHTGTGWCIFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYD 335
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 336 EAAMAIASLNGYRLGDRVLQVSF 358
Score = 39.7 bits (91), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 17/78 (21%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 116 IHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGMDGQEL 175
+ + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G++GQ+
Sbjct 139 LPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGLNGQKP 198
Query 176 LGQKVAVSWAFVRQPEKR 193
G ++ F P ++
Sbjct 199 SGATEPITVKFANNPSQK 216
Score = 39.3 bits (90), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
++ + + +E+ F + G I++ + D+ +G GY FV Y +A+ AI +
Sbjct 48 LIVNYLPQNMTQEEFRSLFGSIGEIESCKLVRDKITGQSLGYGFVNYIDPKDAEKAINTL 107
Query 171 DGQELLGQKVAVSWA 185
+G L + + VS+A
Sbjct 108 NGLRLQTKTIKVSYA 122
> xla:386602 elavl4, HuD, elrD, elrD1, elrD2; ELAV (embryonic
lethal, abnormal vision)-like 4 (Hu antigen D); K13208 ELAV
like protein 2/3/4
Length=400
Score = 53.9 bits (128), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 42/83 (50%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P + GW + N+ ++ E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 310 PGHTGTGWCIFVYNLSPDSDESVLWQLFGPFGAVNNVKVIRDFNTNKCKGFGFVTMTNYD 369
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 370 EAAMAIASLNGYRLGDRVLQVSF 392
Score = 38.9 bits (89), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 17/83 (20%), Positives = 44/83 (53%), Gaps = 0/83 (0%)
Query 111 VVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYDEAQAAIEGM 170
+ + + + ++++ + F +G I + +D+ +G +G F+ ++ EA+ AI+G+
Sbjct 168 LYVSGLPKTMTQKELEQLFSQYGRIITSRILVDQVTGVSRGVGFIRFDKRIEAEEAIKGL 227
Query 171 DGQELLGQKVAVSWAFVRQPEKR 193
+GQ+ G ++ F P ++
Sbjct 228 NGQKPSGAAEPITVKFANNPSQK 250
> xla:446311 eif3s4, MGC83369, eIF3g-A, eif3s4-A; eukaryotic translation
initiation factor 3, subunit 4 delta, 44kDa; K03248
translation initiation factor 3 subunit G
Length=308
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 30/95 (31%), Positives = 51/95 (53%), Gaps = 1/95 (1%)
Query 91 GKRRGKDKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVK 150
G RRG+ P R+ + + TN+ E+ +E D+ E F FG+I +++ D+ +G K
Sbjct 210 GSRRGESMQPN-RRADDNATIRVTNLSEDTRETDLQELFRPFGSISRIYLAKDKTTGQSK 268
Query 151 GYAFVEYETYDEAQAAIEGMDGQELLGQKVAVSWA 185
G+AF+ + ++A AI G+ G + V WA
Sbjct 269 GFAFISFHRREDAARAIAGVSGFGYDHLILNVEWA 303
> mmu:19826 Rnps1; ribonucleic acid binding protein S1; K14325
RNA-binding protein with serine-rich domain 1
Length=305
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query 90 EGKRRGKDKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDR-RSGF 148
E +R+ + P P + G + ++ I E F +G IK + M ++R
Sbjct 148 EKERKRRSPSPKPTKVHIGRLT------RNVTKDHIMEIFSTYGKIKMIDMPVERMHPHL 201
Query 149 VKGYAFVEYETYDEAQAAIEGMDGQELLGQKVAVS 183
KGYA+VE+E DEA+ A++ MDG ++ GQ++ +
Sbjct 202 SKGYAYVEFENPDEAEKALKHMDGGQIDGQEITAT 236
> hsa:10921 RNPS1, E5.1, MGC117332; RNA binding protein S1, serine-rich
domain; K14325 RNA-binding protein with serine-rich
domain 1
Length=305
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query 90 EGKRRGKDKGPAPARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDR-RSGF 148
E +R+ + P P + G + ++ I E F +G IK + M ++R
Sbjct 148 EKERKRRSPSPKPTKVHIGRLT------RNVTKDHIMEIFSTYGKIKMIDMPVERMHPHL 201
Query 149 VKGYAFVEYETYDEAQAAIEGMDGQELLGQKVAVS 183
KGYA+VE+E DEA+ A++ MDG ++ GQ++ +
Sbjct 202 SKGYAYVEFENPDEAEKALKHMDGGQIDGQEITAT 236
> mmu:15569 Elavl2, Hub, mel-N1; ELAV (embryonic lethal, abnormal
vision, Drosophila)-like 2 (Hu antigen B); K13208 ELAV like
protein 2/3/4
Length=359
Score = 53.5 bits (127), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 41/83 (49%), Gaps = 0/83 (0%)
Query 102 PARSVEGWIVVATNIHEEAQEEDIHEAFDAFGAIKNLHMNLDRRSGFVKGYAFVEYETYD 161
P GW + N+ +A E + + F FGA+ N+ + D + KG+ FV YD
Sbjct 269 PGHPGTGWCIFVYNLAPDADESILWQMFGPFGAVTNVKVIRDFNTNKCKGFGFVTMTNYD 328
Query 162 EAQAAIEGMDGQELLGQKVAVSW 184
EA AI ++G L + + VS+
Sbjct 329 EAAMAIASLNGYRLGDRVLQVSF 351
Lambda K H
0.314 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5608917492
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40