bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2102_orf1
Length=245
Score E
Sequences producing significant alignments: (Bits) Value
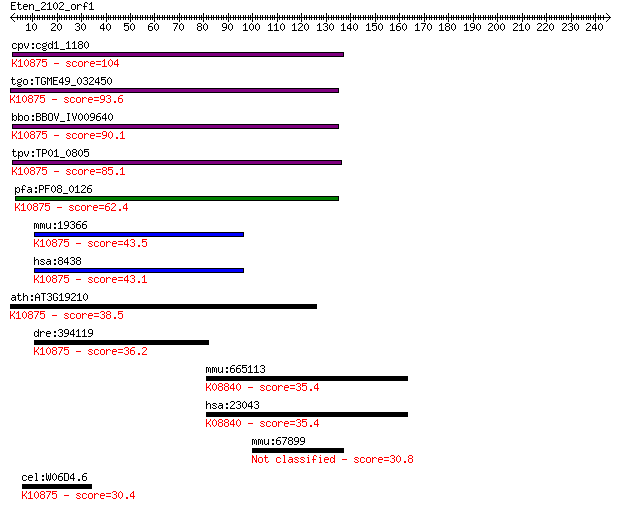
cpv:cgd1_1180 RAD54 like SWI/SNF2 ATpase ; K10875 DNA repair a... 104 3e-22
tgo:TGME49_032450 DNA repair protein RAD54, putative (EC:2.7.1... 93.6 6e-19
bbo:BBOV_IV009640 23.m05735; DNA repair and recombination prot... 90.1 7e-18
tpv:TP01_0805 DNA repair protein Rad54; K10875 DNA repair and ... 85.1 2e-16
pfa:PF08_0126 DNA repair protein rad54, putative; K10875 DNA r... 62.4 1e-09
mmu:19366 Rad54l, RAD54; RAD54 like (S. cerevisiae); K10875 DN... 43.5 8e-04
hsa:8438 RAD54L, HR54, RAD54A, hHR54, hRAD54; RAD54-like (S. c... 43.1 0.001
ath:AT3G19210 ATRAD54 (ARABIDOPSIS HOMOLOG OF RAD54); ATP bind... 38.5 0.021
dre:394119 rad54l, MGC56289, zgc:56289; RAD54-like (S. cerevis... 36.2 0.12
mmu:665113 Tnik, 1500031A17Rik, 4831440I19Rik, AI451411, C5300... 35.4 0.18
hsa:23043 TNIK; TRAF2 and NCK interacting kinase (EC:2.7.11.1)... 35.4 0.20
mmu:67899 Cmc1, 2010110K16Rik, 2010312P05Rik, AI596001; COX as... 30.8 4.5
cel:W06D4.6 rad-54; RADiation sensitivity abnormal/yeast RAD-r... 30.4 6.0
> cpv:cgd1_1180 RAD54 like SWI/SNF2 ATpase ; K10875 DNA repair
and recombination protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=877
Score = 104 bits (259), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 62/141 (43%), Positives = 82/141 (58%), Gaps = 11/141 (7%)
Query 2 LKDSLSTDIVKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFEEDDLLTW 60
LKDS+S D+V+DLF L+E T SDTHDM+QC RC S +PL MV Q G E DDL TW
Sbjct 668 LKDSISADLVRDLFTLKEDTISDTHDMIQCNRCHDS-NGQPLDMVPQ-TTGLE-DDLNTW 724
Query 61 AHHSDLKTVPDEMLLKAIEAAEGD----AADALSSDTHEAPTEIPSHFQPVSFAMSCKVE 116
H+ +PDE+L + + A D H P E+PS FQPVSF M+C++E
Sbjct 725 GHYHSFSEIPDEILSLTLNECSNETNKMAVDMEGEKPH--PLELPSDFQPVSFVMACRIE 782
Query 117 FT-KESEETNSAGAPAPKKVN 136
+E E+T + P +V+
Sbjct 783 LQDQEDEQTKESNGEKPMEVD 803
> tgo:TGME49_032450 DNA repair protein RAD54, putative (EC:2.7.11.1);
K10875 DNA repair and recombination protein RAD54 and
RAD54-like protein [EC:3.6.4.-]
Length=873
Score = 93.6 bits (231), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 71/157 (45%), Positives = 93/157 (59%), Gaps = 29/157 (18%)
Query 1 RLKDSLSTDIVKDLFQLRE-TRSDTHDMLQCQRC-------------------QPSRETE 40
++KDSLST++VKDLF+LRE T SDTHDML+C+RC RETE
Sbjct 669 QIKDSLSTELVKDLFRLREDTLSDTHDMLECRRCGCRAAGQDVENEGENERREAKRRETE 728
Query 41 P--LGMVAQLVDGFEEDDLLTWAHHSDLKTVPDEMLLKAIEAAEGDAADALSSDTHEAPT 98
LG V QL + F+EDDL+TWAHH DL +VPD L +A A D L+ D +
Sbjct 729 EEMLGSVPQLDEDFDEDDLVTWAHHRDLASVPDPCLRQAALA----CVDPLACDRED--E 782
Query 99 EIPSHFQPVSFAMSCKVEFTKESE-ETNSAGAPAPKK 134
E+ S QP+SF MSC++EF E + ++ +A AP K
Sbjct 783 EMISSEQPISFVMSCRIEFKDEGKPQSPAAAAPVCGK 819
> bbo:BBOV_IV009640 23.m05735; DNA repair and recombination protein
RAD54-like (EC:3.6.1.-); K10875 DNA repair and recombination
protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=824
Score = 90.1 bits (222), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 61/141 (43%), Positives = 74/141 (52%), Gaps = 28/141 (19%)
Query 2 LKDSLSTDIVKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFEEDDLLTW 60
+KDSLS + +++LF+ +E T SDTHD L C RC E G V Q D F EDDLLTW
Sbjct 639 IKDSLSGEYLRNLFEFKEDTISDTHDSLSCTRCN-----ETTGHVPQTAD-FVEDDLLTW 692
Query 61 AHHSDLKTVPDEMLLKAIEAAEGDAADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKE 120
AHH+DL T+PD L AI + P + + PVSF MSC VEF KE
Sbjct 693 AHHTDLNTLPDPCLKSAIFE-------------NNPPVQTDEGYTPVSFVMSCLVEF-KE 738
Query 121 SE-------ETNSAGAPAPKK 134
S+ T A P P K
Sbjct 739 SQTPQQRPVNTEPAVKPQPLK 759
> tpv:TP01_0805 DNA repair protein Rad54; K10875 DNA repair and
recombination protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=786
Score = 85.1 bits (209), Expect = 2e-16, Method: Composition-based stats.
Identities = 58/143 (40%), Positives = 78/143 (54%), Gaps = 19/143 (13%)
Query 2 LKDSLSTDIVKDLFQLRETR-SDTHDMLQCQRC--QPSRETE------PLGMVAQLVDGF 52
LKDSLS + +K+LF+ +E SDTHD+++C+RC Q + + P+ V Q D F
Sbjct 626 LKDSLSGEYLKNLFEYKENIISDTHDLIECKRCSNQDGKFVKLIITLGPVMHVPQDKD-F 684
Query 53 EEDDLLTWAHHSDLKTVPDEMLLKAIEAAEGDAADALSSDTHEAPTEIPSHFQPVSFAMS 112
EDDL TWAHHSDL T+PDE L+ A E I F PVSF M+
Sbjct 685 LEDDLNTWAHHSDLTTIPDEYLVSAATKNE--------PQIQYDDKRISDDFVPVSFVMT 736
Query 113 CKVEFTKESEETNSAGAPAPKKV 135
C++EF + EE+ S KK+
Sbjct 737 CRIEF-QNVEESVSVENIGVKKI 758
> pfa:PF08_0126 DNA repair protein rad54, putative; K10875 DNA
repair and recombination protein RAD54 and RAD54-like protein
[EC:3.6.4.-]
Length=1239
Score = 62.4 bits (150), Expect = 1e-09, Method: Composition-based stats.
Identities = 50/182 (27%), Positives = 77/182 (42%), Gaps = 54/182 (29%)
Query 3 KDSLSTDIVKDLFQLR-ETRSDTHDMLQCQRCQPSRETEPLGMVAQL------------- 48
KD +S + VK LF + T S+THD ++C RC + + + VA +
Sbjct 766 KDQMSDENVKKLFNYKMNTVSETHDNIECNRCNLNGVSPNIHKVAHVDSINKNKSKGKND 825
Query 49 ---------------------------VDG---------FEEDDLLTWAHHSDLKTVPDE 72
+DG FEEDD+ TWAHH ++ TVPD
Sbjct 826 NNNNNNNNNSNSNGNGNNYKSDDDKNYIDGEHFAEQLEDFEEDDVNTWAHHQNIDTVPDN 885
Query 73 MLLKAIEAAEGDAADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKESEETNSAGAPAP 132
+L+KA++ DA + +D + P V+F MSCK+E+ + + S A
Sbjct 886 ILIKAVK----DAEEYKKNDMNSMPLLQKLTHDFVTFTMSCKIEYRDDLIKQKSEQAKMR 941
Query 133 KK 134
+K
Sbjct 942 EK 943
> mmu:19366 Rad54l, RAD54; RAD54 like (S. cerevisiae); K10875
DNA repair and recombination protein RAD54 and RAD54-like protein
[EC:3.6.4.-]
Length=747
Score = 43.5 bits (101), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 44/87 (50%), Gaps = 8/87 (9%)
Query 11 VKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFE-EDDLLTWAHHSDLKT 68
+K+LF L E + SDTHD L C+RC +R+ P DG + DL W H +D +
Sbjct 661 LKELFTLDEASLSDTHDRLHCRRCVNNRQVWPPP------DGSDCTSDLAQWNHSTDKRG 714
Query 69 VPDEMLLKAIEAAEGDAADALSSDTHE 95
+ DE+L A +A+ +HE
Sbjct 715 LQDEVLQAAWDASSTAITFVFHQRSHE 741
> hsa:8438 RAD54L, HR54, RAD54A, hHR54, hRAD54; RAD54-like (S.
cerevisiae); K10875 DNA repair and recombination protein RAD54
and RAD54-like protein [EC:3.6.4.-]
Length=747
Score = 43.1 bits (100), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/87 (36%), Positives = 43/87 (49%), Gaps = 8/87 (9%)
Query 11 VKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFE-EDDLLTWAHHSDLKT 68
+K+LF L E + SDTHD L C+RC SR+ P DG + DL W H +D
Sbjct 661 LKELFILDEASLSDTHDRLHCRRCVNSRQIRPPP------DGSDCTSDLAGWNHCTDKWG 714
Query 69 VPDEMLLKAIEAAEGDAADALSSDTHE 95
+ DE+L A +AA +HE
Sbjct 715 LRDEVLQAAWDAASTAITFVFHQRSHE 741
> ath:AT3G19210 ATRAD54 (ARABIDOPSIS HOMOLOG OF RAD54); ATP binding
/ DNA binding / helicase/ nucleic acid binding; K10875
DNA repair and recombination protein RAD54 and RAD54-like protein
[EC:3.6.4.-]
Length=910
Score = 38.5 bits (88), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 42/160 (26%), Positives = 63/160 (39%), Gaps = 48/160 (30%)
Query 1 RLKDSLSTDIVKDLFQLR-ETRSDTHDMLQCQRCQP---------------------SRE 38
R + LST+ ++DLF + RS+ H+ + C RCQ +
Sbjct 693 RQGNLLSTEDLRDLFSFHGDVRSEIHEKMSCSRCQNDASGTENIEEGNENNVDDNACQID 752
Query 39 TEPLGMVAQLVDGFE-------------EDDLLTWAHHSDLKTVPDEMLLKAIEAAEGDA 85
E +G A+ F E+DL +W HH K+VPD +L +A+ GD
Sbjct 753 QEDIGGFAKDAGCFNLLKNSERQVGTPLEEDLGSWGHHFTSKSVPDAIL----QASAGDE 808
Query 86 ADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKESEETN 125
T ++ P+ +S K T ESEE N
Sbjct 809 V------TFVFTNQVDGKLVPIESNVSPK---TVESEEHN 839
> dre:394119 rad54l, MGC56289, zgc:56289; RAD54-like (S. cerevisiae);
K10875 DNA repair and recombination protein RAD54 and
RAD54-like protein [EC:3.6.4.-]
Length=738
Score = 36.2 bits (82), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Query 11 VKDLFQLRE-TRSDTHDMLQCQRCQPSRETEPLGMVAQLVDGFEEDDLLTWAHHSDLKTV 69
+++LF L E T SDTHD +C+RC R+ P D DL W H +D + +
Sbjct 655 LRELFSLNEKTLSDTHDRFRCRRCVNGRQVRP-----PPDDSDCTCDLSNWHHCADKRGL 709
Query 70 PDEMLLKAIEAA 81
D +L + +AA
Sbjct 710 RDPVLQASWDAA 721
> mmu:665113 Tnik, 1500031A17Rik, 4831440I19Rik, AI451411, C530008O15Rik,
C630040K21Rik, KIAA0551, MGC189819, MGC189859; TRAF2
and NCK interacting kinase (EC:2.7.11.1); K08840 TRAF2
and NCK interacting kinase [EC:2.7.11.1]
Length=1352
Score = 35.4 bits (80), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 81 AEGDAADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKESEETNSAGAPAPKKVNHGEA 140
++G A A S HE PT+ S FQ S +VE +++ + S P P ++ +
Sbjct 600 SQGPALTA-SQSVHEQPTKGLSGFQEALNVTSHRVEMPRQNSDPTSENPPLPTRIEKFDR 658
Query 141 CLSEMRVVSSAPPTSPQQTTAL 162
S +R PP PQ+TT++
Sbjct 659 S-SWLRQEEDIPPKVPQRTTSI 679
> hsa:23043 TNIK; TRAF2 and NCK interacting kinase (EC:2.7.11.1);
K08840 TRAF2 and NCK interacting kinase [EC:2.7.11.1]
Length=1352
Score = 35.4 bits (80), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 40/82 (48%), Gaps = 2/82 (2%)
Query 81 AEGDAADALSSDTHEAPTEIPSHFQPVSFAMSCKVEFTKESEETNSAGAPAPKKVNHGEA 140
++G A A S HE PT+ S FQ S +VE +++ + S P P ++ +
Sbjct 600 SQGPALTA-SQSVHEQPTKGLSGFQEALNVTSHRVEMPRQNSDPTSENPPLPTRIEKFDR 658
Query 141 CLSEMRVVSSAPPTSPQQTTAL 162
S +R PP PQ+TT++
Sbjct 659 S-SWLRQEEDIPPKVPQRTTSI 679
> mmu:67899 Cmc1, 2010110K16Rik, 2010312P05Rik, AI596001; COX
assembly mitochondrial protein homolog (S. cerevisiae)
Length=106
Score = 30.8 bits (68), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 11/37 (29%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 100 IPSHFQPVSFAMSCKVEFTKESEETNSAGAPAPKKVN 136
+ +++ +F CK+E+ KE EE G P K++
Sbjct 64 LTAYYNDPAFYEECKLEYLKEREEFRKTGVPTKKRLQ 100
> cel:W06D4.6 rad-54; RADiation sensitivity abnormal/yeast RAD-related
family member (rad-54); K10875 DNA repair and recombination
protein RAD54 and RAD54-like protein [EC:3.6.4.-]
Length=818
Score = 30.4 bits (67), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 22/29 (75%), Gaps = 1/29 (3%)
Query 6 LSTDIVKDLFQLRET-RSDTHDMLQCQRC 33
S++ +++LF+L T SDTH+ L+C+RC
Sbjct 707 FSSEQLRELFKLESTVASDTHEKLKCKRC 735
Lambda K H
0.308 0.124 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8636871780
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40