bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2090_orf3
Length=414
Score E
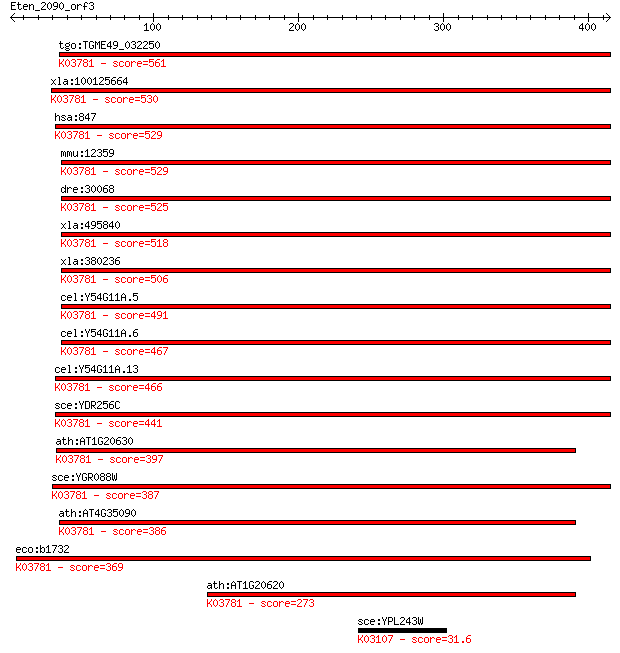
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781 c... 561 2e-159
xla:100125664 hypothetical protein LOC100125664; K03781 catala... 530 6e-150
hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03... 529 7e-150
mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.1... 529 9e-150
dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6); K0... 525 9e-149
xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 cata... 518 2e-146
xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catal... 506 5e-143
cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781 ca... 491 2e-138
cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781 ca... 467 4e-131
cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781 c... 466 9e-131
sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide in... 441 3e-123
ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);... 397 6e-110
sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase... 387 4e-107
ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);... 386 9e-107
eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (ca... 369 2e-101
ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catala... 273 1e-72
sce:YPL243W SRP68; Core component of the signal recognition pa... 31.6 5.3
> tgo:TGME49_032250 peroxisomal catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=502
Score = 561 bits (1445), Expect = 2e-159, Method: Compositional matrix adjust.
Identities = 255/381 (66%), Positives = 310/381 (81%), Gaps = 1/381 (0%)
Query 35 PVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGV 94
PV+TTS G PV DNQNS+TAGPYG +LS+FHL+DKLAHFDRERIPERVVHAKG GA G
Sbjct 14 PVITTSAGNPVDDNQNSVTAGPYGPAILSNFHLIDKLAHFDRERIPERVVHAKGGGAFGY 73
Query 95 FKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWD 154
F+VTHDIT++CKA++FE +GK T VF RFSTV GE GSAD+ RDPRGFA+KFY+ G WD
Sbjct 74 FEVTHDITRFCKAKLFEKIGKRTPVFARFSTVAGESGSADTRRDPRGFALKFYTEEGNWD 133
Query 155 LVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRG 214
+VGNNTP+FF+RD +KFPDFIH+QKR+PQT+L DPNM WDF SL PE++HQ T L++DRG
Sbjct 134 MVGNNTPIFFVRDAIKFPDFIHTQKRHPQTHLHDPNMVWDFFSLVPESVHQVTFLYTDRG 193
Query 215 TPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRD 274
TPDG+RHM+GY SH FK +N + Y K+H KTNQ IKNL+RQRA++L EDPDYA RD
Sbjct 194 TPDGFRHMNGYGSHTFKFINKDNEAFYVKWHFKTNQGIKNLNRQRAKELESEDPDYAVRD 253
Query 275 LFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKN 334
LFNAI FPSWT IQVMP+ +A YK+NV DVTKV P D+PL+PVG+L L +NP+N
Sbjct 254 LFNAIAKREFPSWTFCIQVMPLKDAETYKWNVFDVTKVWPHGDYPLIPVGRLVLDRNPEN 313
Query 335 YFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRA 394
YFQ+VEQ AF+P +++PGIEPSED++LQGR+F+Y DT RHRLG N+ QIPVNRP
Sbjct 314 YFQDVEQAAFAPAHMVPGIEPSEDRMLQGRMFSYIDTHRHRLGANYHQIPVNRPWNARGG 373
Query 395 IH-IRDGPMCVNGNYGALPNY 414
+ +RDGPMCV+GN G+ NY
Sbjct 374 DYSVRDGPMCVDGNKGSQLNY 394
> xla:100125664 hypothetical protein LOC100125664; K03781 catalase
[EC:1.11.1.6]
Length=503
Score = 530 bits (1364), Expect = 6e-150, Method: Compositional matrix adjust.
Identities = 248/386 (64%), Positives = 294/386 (76%), Gaps = 0/386 (0%)
Query 29 MEEKECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKG 88
M ++ P+LTT G P GD N LTAGP G +L+ D D++AHFDRERIPERVVHAKG
Sbjct 1 MAQQTHPILTTGAGIPTGDKLNVLTAGPRGPMLMQDVVFTDEMAHFDRERIPERVVHAKG 60
Query 89 AGAHGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYS 148
AGA G F+VTHDITKYC+A+VFE +GK T+V VRFSTV GE GSAD+ RDPRGFAVKFY+
Sbjct 61 AGAFGYFEVTHDITKYCRAKVFEKVGKRTDVAVRFSTVAGEAGSADTVRDPRGFAVKFYT 120
Query 149 AAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTI 208
G+WDLVGNN P+FF+RDP+ FP FIHSQKRNPQT+L DP+ WDF SL PETLHQ T
Sbjct 121 EEGIWDLVGNNIPIFFLRDPMMFPSFIHSQKRNPQTHLKDPDTVWDFWSLRPETLHQVTF 180
Query 209 LFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDP 268
LFSDRG PDG+RHM+GY SH FKLVN+EG Y KFH KT+Q IKNLS + A +L DP
Sbjct 181 LFSDRGIPDGHRHMNGYGSHTFKLVNAEGNAVYCKFHYKTDQGIKNLSVEEADRLVVSDP 240
Query 269 DYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTL 328
DY RDLF AI NFPSWT+YIQVM EA K +N D+TKV P KD+PL+P+G+L L
Sbjct 241 DYGIRDLFQAIAKKNFPSWTMYIQVMTFEEAEKCPFNPFDLTKVWPHKDYPLIPIGKLVL 300
Query 329 TKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRP 388
+NP+NYF EVEQ+AF P ++ PGIE S DK+LQGRLF+YPDT R+RLGPN+ +PVN P
Sbjct 301 NRNPENYFAEVEQIAFDPSHMPPGIEASPDKMLQGRLFSYPDTHRYRLGPNYLHLPVNCP 360
Query 389 RCPLRAIHIRDGPMCVNGNYGALPNY 414
R A + RDGPMC+ +PNY
Sbjct 361 RGVKVAHYQRDGPMCMFNTPSHMPNY 386
> hsa:847 CAT, MGC138422, MGC138424; catalase (EC:1.11.1.6); K03781
catalase [EC:1.11.1.6]
Length=527
Score = 529 bits (1363), Expect = 7e-150, Method: Compositional matrix adjust.
Identities = 252/386 (65%), Positives = 295/386 (76%), Gaps = 6/386 (1%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
++ VLTT G PVGD N +T GP G +L+ D D++AHFDRERIPERVVHAKGAGA
Sbjct 22 QKADVLTTGAGNPVGDKLNVITVGPRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGA 81
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
G F+VTHDITKY KA+VFE +GK+T + VRFSTV GE GSAD+ RDPRGFAVKFY+ G
Sbjct 82 FGYFEVTHDITKYSKAKVFEHIGKKTPIAVRFSTVAGESGSADTVRDPRGFAVKFYTEDG 141
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFS 211
WDLVGNNTP+FFIRDP+ FP FIHSQKRNPQT+L DP+M WDF SL PE+LHQ + LFS
Sbjct 142 NWDLVGNNTPIFFIRDPILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLHQVSFLFS 201
Query 212 DRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYA 271
DRG PDG+RHM+GY SH FKLVN+ G Y KFH KT+Q IKNLS + A +L+ EDPDY
Sbjct 202 DRGIPDGHRHMNGYGSHTFKLVNANGEAVYCKFHYKTDQGIKNLSVEDAARLSQEDPDYG 261
Query 272 TRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKN 331
RDLFNAI +G +PSWT YIQVM +A + +N D+TKV P KD+PL+PVG+L L +N
Sbjct 262 IRDLFNAIATGKYPSWTFYIQVMTFNQAETFPFNPFDLTKVWPHKDYPLIPVGKLVLNRN 321
Query 332 PKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCP 391
P NYF EVEQ+AF P N+ PGIE S DK+LQGRLFAYPDT RHRLGPN+ IPVN CP
Sbjct 322 PVNYFAEVEQIAFDPSNMPPGIEASPDKMLQGRLFAYPDTHRHRLGPNYLHIPVN---CP 378
Query 392 LRAI---HIRDGPMCVNGNYGALPNY 414
RA + RDGPMC+ N G PNY
Sbjct 379 YRARVANYQRDGPMCMQDNQGGAPNY 404
> mmu:12359 Cat, 2210418N07, Cas-1, Cas1, Cs-1; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=527
Score = 529 bits (1362), Expect = 9e-150, Method: Compositional matrix adjust.
Identities = 254/382 (66%), Positives = 294/382 (76%), Gaps = 6/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
VLTT G P+GD N +TAG G +L+ D D++AHFDRERIPERVVHAKGAGA G F
Sbjct 26 VLTTGGGNPIGDKLNIMTAGSRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGAFGYF 85
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDIT+Y KA+VFE +GK T + VRFSTV GE GSAD+ RDPRGFAVKFY+ G WDL
Sbjct 86 EVTHDITRYSKAKVFEHIGKRTPIAVRFSTVTGESGSADTVRDPRGFAVKFYTEDGNWDL 145
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTP+FFIRD + FP FIHSQKRNPQT+L DP+M WDF SL PE+LHQ + LFSDRG
Sbjct 146 VGNNTPIFFIRDAILFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLHQVSFLFSDRGI 205
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDG+RHM+GY SH FKLVN++G Y KFH KT+Q IKNL A +LA EDPDY RDL
Sbjct 206 PDGHRHMNGYGSHTFKLVNADGEAVYCKFHYKTDQGIKNLPVGEAGRLAQEDPDYGLRDL 265
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
FNAI +GN+PSWT YIQVM EA + +N D+TKV P KD+PL+PVG+L L KNP NY
Sbjct 266 FNAIANGNYPSWTFYIQVMTFKEAETFPFNPFDLTKVWPHKDYPLIPVGKLVLNKNPVNY 325
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ+AF P N+ PGIEPS DK+LQGRLFAYPDT RHRLGPN+ QIPVN CP RA
Sbjct 326 FAEVEQMAFDPSNMPPGIEPSPDKMLQGRLFAYPDTHRHRLGPNYLQIPVN---CPYRAR 382
Query 396 ---HIRDGPMCVNGNYGALPNY 414
+ RDGPMC++ N G PNY
Sbjct 383 VANYQRDGPMCMHDNQGGAPNY 404
> dre:30068 cat, fb68a12, wu:fb68a12; catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=526
Score = 525 bits (1353), Expect = 9e-149, Method: Compositional matrix adjust.
Identities = 248/382 (64%), Positives = 294/382 (76%), Gaps = 6/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
VLTT G P+GD N++TAGP G +L+ D D++AHFDRERIPERVVHAKGAGA G F
Sbjct 26 VLTTGAGVPIGDKLNAMTAGPRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGAFGYF 85
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDIT+Y KA+VFE +GK T + VRFSTV GE GS+D+ RDPRGFAVKFY+ G WDL
Sbjct 86 EVTHDITRYSKAKVFEHVGKTTPIAVRFSTVAGEAGSSDTVRDPRGFAVKFYTDEGNWDL 145
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
GNNTP+FFIRD L FP FIHSQKRNPQT+L DP+M WDF SL PE+LHQ + LFSDRG
Sbjct 146 TGNNTPIFFIRDTLLFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLHQVSFLFSDRGI 205
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDGYRHM+GY SH FKLVN++G Y KFH KTNQ IKN+ + A +LA DPDY+ RDL
Sbjct 206 PDGYRHMNGYGSHTFKLVNAQGQPVYCKFHYKTNQGIKNIPVEEADRLAATDPDYSIRDL 265
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
+NAI +GNFPSWT YIQVM +A +K+N D+TKV K+FPL+PVG+ L +NP NY
Sbjct 266 YNAIANGNFPSWTFYIQVMTFEQAENWKWNPFDLTKVWSHKEFPLIPVGRFVLNRNPVNY 325
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLR-- 393
F EVEQ+AF P N+ PGIEPS DK+LQGRLF+YPDT RHRLG N+ Q+PVN CP R
Sbjct 326 FAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGANYLQLPVN---CPYRTR 382
Query 394 -AIHIRDGPMCVNGNYGALPNY 414
A + RDGPMC++ N G PNY
Sbjct 383 VANYQRDGPMCMHDNQGGAPNY 404
> xla:495840 cat, MGC116544; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 518 bits (1334), Expect = 2e-146, Method: Compositional matrix adjust.
Identities = 240/379 (63%), Positives = 288/379 (75%), Gaps = 0/379 (0%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
VLTT G PVGD N LT GP G +L+ D D++AHFDRERIPERVVHAKGAGA G F
Sbjct 26 VLTTGAGHPVGDKLNLLTVGPRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGAFGYF 85
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDIT+YCKA+VFE +GK T V RFSTV GE GS D+ RDPRGFAVK Y+ G WDL
Sbjct 86 EVTHDITQYCKAKVFEKVGKRTPVAARFSTVAGEAGSPDTIRDPRGFAVKMYTEDGNWDL 145
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
GNNTP+FFIRD + FP F+HSQ+RNPQT++ DPNM WDF +L PE+LHQ + LFSDRG
Sbjct 146 TGNNTPIFFIRDAILFPSFVHSQERNPQTHMKDPNMVWDFWTLRPESLHQVSFLFSDRGI 205
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDG+RHM+GY SH FKLV+ + Y KFH KT+Q I+NL+ +RA+QLA DPDY RDL
Sbjct 206 PDGHRHMNGYGSHTFKLVSCKDEAVYCKFHFKTDQGIRNLTVERAEQLAASDPDYGIRDL 265
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
+ AI +GN+PSWTLYIQ M +A K+ +N DVTKV P D+PL+PVG++ L++NPKNY
Sbjct 266 YEAIAAGNYPSWTLYIQTMTFQQAEKFPFNPFDVTKVWPHGDYPLIPVGKMVLSQNPKNY 325
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ+AF P N+ PGIEPS DK+LQGRLF+YPDT RHRLG N+ Q+P+N P A
Sbjct 326 FAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGTNYLQLPINCPYKVRVAN 385
Query 396 HIRDGPMCVNGNYGALPNY 414
+ RDGPMC N G PNY
Sbjct 386 YQRDGPMCFTDNQGGAPNY 404
> xla:380236 cat, MGC64369; catalase (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=528
Score = 506 bits (1304), Expect = 5e-143, Method: Compositional matrix adjust.
Identities = 239/382 (62%), Positives = 285/382 (74%), Gaps = 6/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
VLTT G P+ D N LT GP G +L+ D D++AHFDRERIPERVVHAKGAGA G
Sbjct 26 VLTTGGGNPISDKLNLLTVGPRGPLLVQDVVFTDEMAHFDRERIPERVVHAKGAGAFGYC 85
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDITKY KA+VFE +GK T + VRFSTV GE GS+D+ RDPRGFAVK Y+ G WDL
Sbjct 86 EVTHDITKYSKAKVFENIGKRTPIAVRFSTVAGEAGSSDTVRDPRGFAVKMYTEDGNWDL 145
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
GNNTPVFFIRD + FP FIHSQKRNPQT+L DP+M WDF SL PE+LHQ + LFSDRG
Sbjct 146 TGNNTPVFFIRDAMLFPSFIHSQKRNPQTHLKDPDMVWDFWSLRPESLHQVSFLFSDRGI 205
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDG+RHM+GY SH FKLVN++ Y KFH KT+Q I+NL+ A +LA DPDY DL
Sbjct 206 PDGHRHMNGYGSHTFKLVNAKDEAVYCKFHYKTDQCIQNLTVDEANRLAASDPDYGIHDL 265
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
+ AI +GN+PSW+ YIQVM +A ++K+N D+TK+ P D+PL+PVG+L L +NP NY
Sbjct 266 YEAITTGNYPSWSFYIQVMTFEQAERFKFNPFDLTKIWPHGDYPLIPVGKLVLNRNPTNY 325
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLR-- 393
F EVEQ+AF P N+ PGIEPS DK+LQGRLF+YPDT RHRLGPN+ Q+PVN CP R
Sbjct 326 FAEVEQLAFDPSNMPPGIEPSPDKMLQGRLFSYPDTHRHRLGPNYLQLPVN---CPYRTR 382
Query 394 -AIHIRDGPMCVNGNYGALPNY 414
A + RD PMC N G PNY
Sbjct 383 VANYQRDRPMCFTDNQGGAPNY 404
> cel:Y54G11A.5 ctl-2; CaTaLase family member (ctl-2); K03781
catalase [EC:1.11.1.6]
Length=500
Score = 491 bits (1264), Expect = 2e-138, Method: Compositional matrix adjust.
Identities = 231/382 (60%), Positives = 288/382 (75%), Gaps = 7/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
V+TTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKGAGAHG F
Sbjct 22 VITTSNGAPIYSKTAVLTAGRRGPMLMQDVVYMDEMAHFDRERIPERVVHAKGAGAHGYF 81
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDI+KYCKA++F +GK+T + +RFSTVGGE GSAD+ARDPRGFA+KFY+ G WDL
Sbjct 82 EVTHDISKYCKADIFNKVGKQTPLLIRFSTVGGESGSADTARDPRGFAIKFYTEEGNWDL 141
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTP+FFIRDP+ FP+FIH+QKRNPQT+L DPNM +DF PE LHQ LFSDRG
Sbjct 142 VGNNTPIFFIRDPIHFPNFIHTQKRNPQTHLKDPNMIFDFWLHRPEALHQVMFLFSDRGL 201
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDGYRHM+GY SH FK+VN +G Y KFH K Q +KNL+ ++A QLA DPDY+ RDL
Sbjct 202 PDGYRHMNGYGSHTFKMVNKDGKAIYVKFHFKPTQGVKNLTVEKAGQLASSDPDYSIRDL 261
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
FNAI+ G+FP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +NP+NY
Sbjct 262 FNAIEKGDFPVWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRNPRNY 321
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRAI 395
F EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP R+
Sbjct 322 FAEVEQSAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHFHRLGPNYIQLPVN---CPYRSR 378
Query 396 H---IRDGPMCVNGNYGALPNY 414
RDG M + A PN+
Sbjct 379 AHNTQRDGAMAYDNQQHA-PNF 399
> cel:Y54G11A.6 ctl-1; CaTaLase family member (ctl-1); K03781
catalase [EC:1.11.1.6]
Length=497
Score = 467 bits (1202), Expect = 4e-131, Method: Compositional matrix adjust.
Identities = 225/382 (58%), Positives = 278/382 (72%), Gaps = 7/382 (1%)
Query 36 VLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGVF 95
V+TTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKGAGAHG F
Sbjct 22 VITTSNGAPIYSKTAVLTAGRRGPMLMQDVVYMDEMAHFDRERIPERVVHAKGAGAHGYF 81
Query 96 KVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWDL 155
+VTHDITKYCKA++F +GK+T + VRFSTV GE GSAD+ RDPRGF++KFY+ G WDL
Sbjct 82 EVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSLKFYTEEGNWDL 141
Query 156 VGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRGT 215
VGNNTP+FFIRD + FP+FIH+ KRNPQT++ DPN +DF PE++HQ L+SDRG
Sbjct 142 VGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIHQVMFLYSDRGI 201
Query 216 PDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDL 275
PDG+R M+GY +H FK+VN EG Y KFH K Q KNL A +LA DPDYA RDL
Sbjct 202 PDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTDAGKLASSDPDYAIRDL 261
Query 276 FNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNY 335
FNAI+S NFP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +N KNY
Sbjct 262 FNAIESRNFPEWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRNVKNY 321
Query 336 FQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCPLRA- 394
F EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP R+
Sbjct 322 FAEVEQAAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHYHRLGPNYIQLPVN---CPYRSR 378
Query 395 IHI--RDGPMCVNGNYGALPNY 414
H RDG M G PNY
Sbjct 379 AHTTQRDGAMAYESQ-GDAPNY 399
> cel:Y54G11A.13 ctl-3; CaTaLase family member (ctl-3); K03781
catalase [EC:1.11.1.6]
Length=512
Score = 466 bits (1198), Expect = 9e-131, Method: Compositional matrix adjust.
Identities = 225/386 (58%), Positives = 279/386 (72%), Gaps = 7/386 (1%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
+E +LTTS GAP+ LTAG G +L+ D +D++AHFDRERIPERVVHAKG GA
Sbjct 33 QEPHLLTTSNGAPIYSKTAVLTAGRRGPMLMQDIVYMDEMAHFDRERIPERVVHAKGGGA 92
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
HG F+VTHDITKYCKA++F +GK+T + VRFSTV GE GSAD+ RDPRGF++KFY+ G
Sbjct 93 HGYFEVTHDITKYCKADMFNKVGKQTPLLVRFSTVAGESGSADTVRDPRGFSLKFYTEEG 152
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFS 211
WDLVGNNTP+FFIRD + FP+FIH+ KRNPQT++ DPN +DF PE++HQ L+S
Sbjct 153 NWDLVGNNTPIFFIRDAIHFPNFIHALKRNPQTHMRDPNALFDFWMNRPESIHQVMFLYS 212
Query 212 DRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYA 271
DRG PDG+R M+GY +H FK+VN EG Y KFH K Q KNL A +LA DPDYA
Sbjct 213 DRGIPDGFRFMNGYGAHTFKMVNKEGNPIYCKFHFKPAQGSKNLDPTDAGKLASSDPDYA 272
Query 272 TRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKN 331
RDLFNAI+S NFP W ++IQVM +A K+++N DVTKV P D+PL+ VG++ L +N
Sbjct 273 IRDLFNAIESRNFPEWKMFIQVMTFEQAEKWEFNPFDVTKVWPHGDYPLIEVGKMVLNRN 332
Query 332 PKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRCP 391
KNYF EVEQ AF P +++PGIE S DK+LQGR+F+Y DT HRLGPN+ Q+PVN CP
Sbjct 333 VKNYFAEVEQAAFCPAHIVPGIEFSPDKMLQGRIFSYTDTHYHRLGPNYIQLPVN---CP 389
Query 392 LRA-IHI--RDGPMCVNGNYGALPNY 414
R+ H RDG M G PNY
Sbjct 390 YRSRAHTTQRDGAMAYESQ-GDAPNY 414
> sce:YDR256C CTA1; Catalase A, breaks down hydrogen peroxide
in the peroxisomal matrix formed by acyl-CoA oxidase (Pox1p)
during fatty acid beta-oxidation (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=515
Score = 441 bits (1133), Expect = 3e-123, Method: Compositional matrix adjust.
Identities = 217/390 (55%), Positives = 273/390 (70%), Gaps = 11/390 (2%)
Query 32 KECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGA 91
+E V+T STG P+ + + G +G +LL D++L+D LAHF+RE IP+R HA G+GA
Sbjct 17 REDRVVTNSTGNPINEPFVTQRIGEHGPLLLQDYNLIDSLAHFNRENIPQRNPHAHGSGA 76
Query 92 HGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAG 151
G F+VT DIT C + +F +GK T+ RFSTVGG+KGSAD+ RDPRGFA KFY+ G
Sbjct 77 FGYFEVTDDITDICGSAMFSKIGKRTKCLTRFSTVGGDKGSADTVRDPRGFATKFYTEEG 136
Query 152 VWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPE---TLHQTTI 208
D V NNTPVFFIRDP KFP FIH+QKRNPQTNL D +MFWDFL+ +PE +HQ I
Sbjct 137 NLDWVYNNTPVFFIRDPSKFPHFIHTQKRNPQTNLRDADMFWDFLT-TPENQVAIHQVMI 195
Query 209 LFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDP 268
LFSDRGTP YR MHGYS H +K N G HY + H+KT+Q IKNL+ + A ++AG +P
Sbjct 196 LFSDRGTPANYRSMHGYSGHTYKWSNKNGDWHYVQVHIKTDQGIKNLTIEEATKIAGSNP 255
Query 269 DYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTL 328
DY +DLF AIQ+GN+PSWT+YIQ M +A K ++V D+TKV PQ FPL VG++ L
Sbjct 256 DYCQQDLFEAIQNGNYPSWTVYIQTMTERDAKKLPFSVFDLTKVWPQGQFPLRRVGKIVL 315
Query 329 TKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRP 388
+NP N+F +VEQ AF+P +P E S D +LQ RLF+Y D R+RLGPNF QIPVN
Sbjct 316 NENPLNFFAQVEQAAFAPSTTVPYQEASADPVLQARLFSYADAHRYRLGPNFHQIPVN-- 373
Query 389 RCPLRAIH----IRDGPMCVNGNYGALPNY 414
CP + IRDGPM VNGN+G+ P Y
Sbjct 374 -CPYASKFFNPAIRDGPMNVNGNFGSEPTY 402
> ath:AT1G20630 CAT1; CAT1 (CATALASE 1); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=492
Score = 397 bits (1019), Expect = 6e-110, Method: Compositional matrix adjust.
Identities = 189/358 (52%), Positives = 252/358 (70%), Gaps = 0/358 (0%)
Query 33 ECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAH 92
+ P TT++GAPV +N +SLT G G +LL D+HLL+KLA+FDRERIPERVVHA+GA A
Sbjct 13 DSPFFTTNSGAPVWNNNSSLTVGTRGPILLEDYHLLEKLANFDRERIPERVVHARGASAK 72
Query 93 GVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGV 152
G F+VTHDIT+ A+ G G +T V VRFSTV E+GS ++ RDPRGFAVKFY+ G
Sbjct 73 GFFEVTHDITQLTSADFLRGPGVQTPVIVRFSTVIHERGSPETLRDPRGFAVKFYTREGN 132
Query 153 WDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSD 212
+DLVGNN PVFF+RD +KFPD +H+ K NP++++ + DF S PE+LH + LF D
Sbjct 133 FDLVGNNFPVFFVRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLHMFSFLFDD 192
Query 213 RGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYAT 272
G P YRHM G + + L+N G HY KFH K IK LS + A ++ G + +AT
Sbjct 193 LGIPQDYRHMEGAGVNTYMLINKAGKAHYVKFHWKPTCGIKCLSDEEAIRVGGANHSHAT 252
Query 273 RDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNP 332
+DL+++I +GN+P W L++QVM A K+ ++ LDVTK+ P+ PL PVG+L L KN
Sbjct 253 KDLYDSIAAGNYPQWNLFVQVMDPAHEDKFDFDPLDVTKIWPEDILPLQPVGRLVLNKNI 312
Query 333 KNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRC 390
N+F E EQ+AF P ++PGI S+DKLLQ R+F+Y D+QRHRLGPN+ Q+PVN P+C
Sbjct 313 DNFFNENEQIAFCPALVVPGIHYSDDKLLQTRIFSYADSQRHRLGPNYLQLPVNAPKC 370
> sce:YGR088W CTT1, SPS101; Ctt1p (EC:1.11.1.6); K03781 catalase
[EC:1.11.1.6]
Length=562
Score = 387 bits (994), Expect = 4e-107, Method: Compositional matrix adjust.
Identities = 197/408 (48%), Positives = 266/408 (65%), Gaps = 24/408 (5%)
Query 30 EEKECPVLTTSTGAPVGDN-QNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKG 88
EEK+ V + G P + S + P G +LL DFHLL+ +A FDRER+PERVVHAKG
Sbjct 8 EEKQEKVYSLQNGFPYSHHPYASQYSRPDGPILLQDFHLLENIASFDRERVPERVVHAKG 67
Query 89 AGAHGVFKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYS 148
G F++T ++ A ++ +G + VRFSTVGGE G+ D+ARDPRG + KFY+
Sbjct 68 GGCRLEFELTDSLSDITYAAPYQNVGYKCPGLVRFSTVGGESGTPDTARDPRGVSFKFYT 127
Query 149 AAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNL---PDPNMFWDFLSLSPETLHQ 205
G D V NNTPVFF+RD +KFP FIHSQKR+PQ++L D ++WD+L+L+PE++HQ
Sbjct 128 EWGNHDWVFNNTPVFFLRDAIKFPVFIHSQKRDPQSHLNQFQDTTIYWDYLTLNPESIHQ 187
Query 206 TTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAG 265
T +F DRGTP + M+ YS H+F +VN EG Y +FH+ ++ + L+ +A +L+G
Sbjct 188 ITYMFGDRGTPASWASMNAYSGHSFIMVNKEGKDTYVQFHVLSDTGFETLTGDKAAELSG 247
Query 266 EDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQ 325
PDY LF +Q+G P + Y+Q M +A K++Y+V D+TK+ P K+FPL G
Sbjct 248 SHPDYNQAKLFTQLQNGEKPKFNCYVQTMTPEQATKFRYSVNDLTKIWPHKEFPLRKFGT 307
Query 326 LTLTKNPKNYFQEVEQVAFSPGN-LIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIP 384
+TLT+N NYFQE+EQVAFSP N IPGI+PS D +LQ RLF+YPDTQRHRLG N+ Q+P
Sbjct 308 ITLTENVDNYFQEIEQVAFSPTNTCIPGIKPSNDSVLQARLFSYPDTQRHRLGANYQQLP 367
Query 385 VNRPR-----------------CPLRAIHI-RDGPMCVNGNYGALPNY 414
VNRPR CP +A++ RDGPM N+G PNY
Sbjct 368 VNRPRNLGCPYSKGDSQYTAEQCPFKAVNFQRDGPMSYY-NFGPEPNY 414
> ath:AT4G35090 CAT2; CAT2 (CATALASE 2); catalase (EC:1.11.1.6);
K03781 catalase [EC:1.11.1.6]
Length=474
Score = 386 bits (991), Expect = 9e-107, Method: Compositional matrix adjust.
Identities = 184/356 (51%), Positives = 250/356 (70%), Gaps = 0/356 (0%)
Query 35 PVLTTSTGAPVGDNQNSLTAGPYGSVLLSDFHLLDKLAHFDRERIPERVVHAKGAGAHGV 94
P TT++GAPV +N +S+T GP G +LL D+HL++KLA+FDRERIPERVVHA+GA A G
Sbjct 15 PFFTTNSGAPVWNNNSSMTVGPRGPILLEDYHLVEKLANFDRERIPERVVHARGASAKGF 74
Query 95 FKVTHDITKYCKAEVFEGLGKETEVFVRFSTVGGEKGSADSARDPRGFAVKFYSAAGVWD 154
F+VTHDI+ A+ G +T V VRFSTV E+GS ++ RDPRGFAVKFY+ G +D
Sbjct 75 FEVTHDISNLTCADFLRAPGVQTPVIVRFSTVIHERGSPETLRDPRGFAVKFYTREGNFD 134
Query 155 LVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFLSLSPETLHQTTILFSDRG 214
LVGNN PVFFIRD +KFPD +H+ K NP++++ + DF S PE+L+ T LF D G
Sbjct 135 LVGNNFPVFFIRDGMKFPDMVHALKPNPKSHIQENWRILDFFSHHPESLNMFTFLFDDIG 194
Query 215 TPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRD 274
P YRHM G + + L+N G HY KFH K +K+L + A ++ G + +AT+D
Sbjct 195 IPQDYRHMDGSGVNTYMLINKAGKAHYVKFHWKPTCGVKSLLEEDAIRVGGTNHSHATQD 254
Query 275 LFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKN 334
L+++I +GN+P W L+IQ++ A+ K+ ++ LDVTK P+ PL PVG++ L KN N
Sbjct 255 LYDSIAAGNYPEWKLFIQIIDPADEDKFDFDPLDVTKTWPEDILPLQPVGRMVLNKNIDN 314
Query 335 YFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRLGPNFDQIPVNRPRC 390
+F E EQ+AF P ++PGI S+DKLLQ R+F+Y DTQRHRLGPN+ Q+PVN P+C
Sbjct 315 FFAENEQLAFCPAIIVPGIHYSDDKLLQTRVFSYADTQRHRLGPNYLQLPVNAPKC 370
> eco:b1732 katE, ECK1730, JW1721; hydroperoxidase HPII(III) (catalase)
(EC:1.11.1.6); K03781 catalase [EC:1.11.1.6]
Length=753
Score = 369 bits (946), Expect = 2e-101, Method: Compositional matrix adjust.
Identities = 184/401 (45%), Positives = 253/401 (63%), Gaps = 6/401 (1%)
Query 5 KPLTSRQTSAPALNSTAGHAFSSTMEEKECPVLTTSTGAPVGDNQNSLTAGPYGSVLLSD 64
+P AP + ++ + E LTT+ G + D+QNSL AG G LL D
Sbjct 48 QPTAPGSLKAPDTRNEKLNSLEDVRKGSENYALTTNQGVRIADDQNSLRAGSRGPTLLED 107
Query 65 FHLLDKLAHFDRERIPERVVHAKGAGAHGVFKVTHDITKYCKAEVFEGLGKETEVFVRFS 124
F L +K+ HFD ERIPER+VHA+G+ AHG F+ ++ KA+ K T VFVRFS
Sbjct 108 FILREKITHFDHERIPERIVHARGSAAHGYFQPYKSLSDITKADFLSDPNKITPVFVRFS 167
Query 125 TVGGEKGSADSARDPRGFAVKFYSAAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQT 184
TV G GSAD+ RD RGFA KFY+ G++DLVGNNTP+FFI+D KFPDF+H+ K P
Sbjct 168 TVQGGAGSADTVRDIRGFATKFYTEEGIFDLVGNNTPIFFIQDAHKFPDFVHAVKPEPHW 227
Query 185 NLPDP----NMFWDFLSLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVH 240
+P + FWD++SL PETLH SDRG P YR M G+ H F+L+N+EG
Sbjct 228 AIPQGQSAHDTFWDYVSLQPETLHNVMWAMSDRGIPRSYRTMEGFGIHTFRLINAEGKAT 287
Query 241 YAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAP 300
+ +FH K +L AQ+L G DPD+ R+L+ AI++G+FP + L Q++P +
Sbjct 288 FVRFHWKPLAGKASLVWDEAQKLTGRDPDFHRRELWEAIEAGDFPEYELGFQLIPEEDEF 347
Query 301 KYKYNVLDVTKVLPQKDFPLLPVGQLTLTKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKL 360
K+ +++LD TK++P++ P+ VG++ L +NP N+F E EQ AF PG+++PG++ + D L
Sbjct 348 KFDFDLLDPTKLIPEELVPVQRVGKMVLNRNPDNFFAENEQAAFHPGHIVPGLDFTNDPL 407
Query 361 LQGRLFAYPDTQRHRL-GPNFDQIPVNRPRCPLRAIHIRDG 400
LQGRLF+Y DTQ RL GPNF +IP+NRP CP RDG
Sbjct 408 LQGRLFSYTDTQISRLGGPNFHEIPINRPTCPYHNFQ-RDG 447
> ath:AT1G20620 CAT3; CAT3 (CATALASE 3); catalase; K03781 catalase
[EC:1.11.1.6]
Length=377
Score = 273 bits (697), Expect = 1e-72, Method: Compositional matrix adjust.
Identities = 130/254 (51%), Positives = 175/254 (68%), Gaps = 0/254 (0%)
Query 137 RDPRGFAVKFYSAAGVWDLVGNNTPVFFIRDPLKFPDFIHSQKRNPQTNLPDPNMFWDFL 196
RD RGFAVKFY+ G +DLVGNNTPVFFIRD ++FPD +H+ K NP+TN+ + D++
Sbjct 2 RDIRGFAVKFYTREGNFDLVGNNTPVFFIRDGIQFPDVVHALKPNPKTNIQEYWRILDYM 61
Query 197 SLSPETLHQTTILFSDRGTPDGYRHMHGYSSHAFKLVNSEGVVHYAKFHLKTNQKIKNLS 256
S PE+L +F D G P YRHM G+ H + L+ G V + KFH K IKNL+
Sbjct 62 SHLPESLLTWCWMFDDVGIPQDYRHMEGFGVHTYTLIAKSGKVLFVKFHWKPTCGIKNLT 121
Query 257 RQRAQQLAGEDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAPKYKYNVLDVTKVLPQK 316
+ A+ + G + +AT+DL +AI SGN+P W L+IQ M A+ K+ ++ LDVTK+ P+
Sbjct 122 DEEAKVVGGANHSHATKDLHDAIASGNYPEWKLFIQTMDPADEDKFDFDPLDVTKIWPED 181
Query 317 DFPLLPVGQLTLTKNPKNYFQEVEQVAFSPGNLIPGIEPSEDKLLQGRLFAYPDTQRHRL 376
PL PVG+L L + N+F E EQ+AF+PG ++PGI S+DKLLQ R+FAY DTQRHRL
Sbjct 182 ILPLQPVGRLVLNRTIDNFFNETEQLAFNPGLVVPGIYYSDDKLLQCRIFAYGDTQRHRL 241
Query 377 GPNFDQIPVNRPRC 390
GPN+ Q+PVN P+C
Sbjct 242 GPNYLQLPVNAPKC 255
> sce:YPL243W SRP68; Core component of the signal recognition
particle (SRP) ribonucleoprotein (RNP) complex that functions
in targeting nascent secretory proteins to the endoplasmic
reticulum (ER) membrane; K03107 signal recognition particle
subunit SRP68
Length=599
Score = 31.6 bits (70), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query 241 YAKFHLKTNQKIKNLSRQRAQQLAGEDPDYATRDLFNAIQSGNFPSWTLYIQVMPVAEAP 300
+AK+H K N+K+++L R R + + Y++++ + I S ++ + T I V+ + A
Sbjct 25 FAKYHAKLNKKLQHL-RSRCHLVTKDTKKYSSKNKYGEINSEDYDNKTKLIGVLILLHAE 83
Query 301 K 301
+
Sbjct 84 R 84
Lambda K H
0.318 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 18883150052
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40