bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2086_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
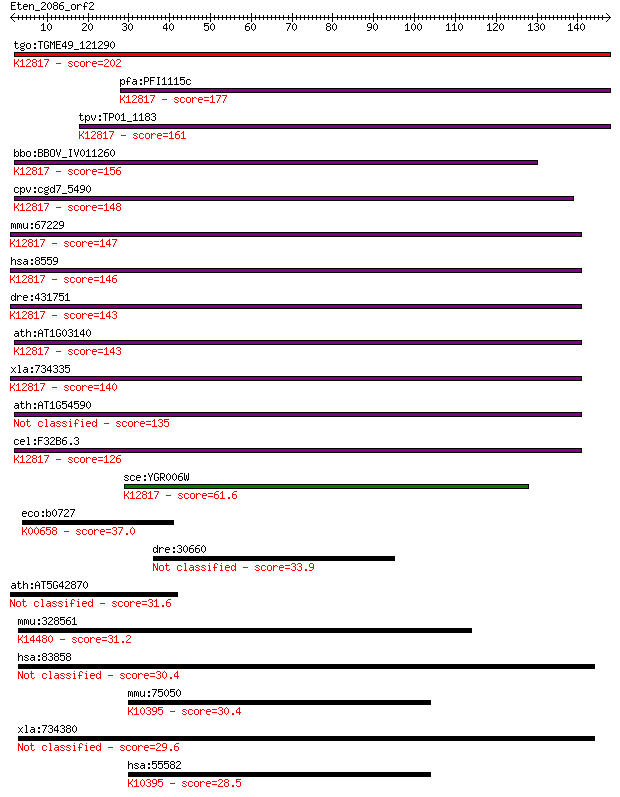
tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K128... 202 2e-52
pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mR... 177 9e-45
tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRN... 161 7e-40
bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K128... 156 2e-38
cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-spli... 148 6e-36
mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing fac... 147 2e-35
hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA proce... 146 2e-35
dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor... 143 1e-34
ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre... 143 2e-34
xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor ... 140 2e-33
ath:AT1G54590 splicing factor Prp18 family protein 135 4e-32
cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing fac... 126 2e-29
sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18 61.6
eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase (... 37.0 0.023
dre:30660 mtnr1al, mel1a, mel1a2, mtnr1a; melatonin receptor t... 33.9 0.18
ath:AT5G42870 lipin family protein 31.6 0.88
mmu:328561 Apol10b, 9130218O11Rik; apolipoprotein L 10b; K1448... 31.2 1.4
hsa:83858 ATAD3B, AAA-TOB3, KIAA1273, TOB3; ATPase family, AAA... 30.4 2.1
mmu:75050 Kif27, 4930517I18Rik; kinesin family member 27; K103... 30.4 2.3
xla:734380 atad3a-a, MGC85169, atad3, atad3-a; ATPase family, ... 29.6 3.2
hsa:55582 KIF27, DKFZp434D0917, RP11-575L7.3; kinesin family m... 28.5 7.4
> tgo:TGME49_121290 pre-mRNA splicing factor 18, putative ; K12817
pre-mRNA-splicing factor 18
Length=373
Score = 202 bits (515), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 91/146 (62%), Positives = 118/146 (80%), Gaps = 0/146 (0%)
Query 2 WEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAK 61
WE EL+ + EE+K + EG+ S+ RQT+KDL+PL+++LR ++LE DIL+KL ++V +
Sbjct 228 WEAELKARPEEKKNSAEGKLMTSLQRQTRKDLKPLLRKLRHKELENDILQKLHIMVQCCE 287
Query 62 ERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQML 121
ERKYR AH ++LLA+GNA WP+GVTMVGIHER GRSKL SQVAHILNDETTRK+IQM
Sbjct 288 ERKYRSAHDTYMLLAIGNAAWPVGVTMVGIHERVGRSKLFSSQVAHILNDETTRKYIQMF 347
Query 122 KRLLSFAQRKFPTDPSQTVLLSVHHV 147
KRL+SF QR++P DPSQT+ LS H+
Sbjct 348 KRLMSFCQRRYPADPSQTISLSTIHI 373
> pfa:PFI1115c pre-mRNA splicing factor, putative; K12817 pre-mRNA-splicing
factor 18
Length=343
Score = 177 bits (450), Expect = 9e-45, Method: Compositional matrix adjust.
Identities = 78/120 (65%), Positives = 101/120 (84%), Gaps = 0/120 (0%)
Query 28 QTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAKERKYREAHGAFVLLAVGNAPWPMGVT 87
QT KDL+PL K+L+Q+ LE DIL+K++ IV+ +E+ ++ AH A++LLA+GNA WPMGVT
Sbjct 224 QTHKDLKPLEKKLKQKTLESDILDKIYNIVSCCQEKNFKAAHDAYMLLAIGNAAWPMGVT 283
Query 88 MVGIHERAGRSKLNVSQVAHILNDETTRKFIQMLKRLLSFAQRKFPTDPSQTVLLSVHHV 147
MVGIHERAGRSK+ S+VAHILNDETTRK+IQM+KRLLSF QRK+ T+PS+ V LS H+
Sbjct 284 MVGIHERAGRSKIYASEVAHILNDETTRKYIQMIKRLLSFCQRKYCTNPSEAVNLSTIHI 343
> tpv:TP01_1183 pre-mRNA splicing factor protein; K12817 pre-mRNA-splicing
factor 18
Length=327
Score = 161 bits (407), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 71/130 (54%), Positives = 101/130 (77%), Gaps = 0/130 (0%)
Query 18 EGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAKERKYREAHGAFVLLAV 77
E ++ ++ QTKKD++PL+K ++ L+ +IL+K+ IVN ++++AH ++LLA+
Sbjct 198 EAKKNEAMLVQTKKDIKPLLKLIKSNKLDQEILDKMEQIVNHCNNGQFKKAHDIYMLLAI 257
Query 78 GNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQMLKRLLSFAQRKFPTDPS 137
GNA WPMGVTMVGIHERAGRSK+ S+VAHILNDETTRK+IQM KRL+SF+Q K+ DPS
Sbjct 258 GNAAWPMGVTMVGIHERAGRSKIFTSEVAHILNDETTRKYIQMFKRLISFSQSKYAKDPS 317
Query 138 QTVLLSVHHV 147
Q + +S +H+
Sbjct 318 QIIQISTNHI 327
> bbo:BBOV_IV011260 23.m06381; pre-mRNA splicing factor 18; K12817
pre-mRNA-splicing factor 18
Length=285
Score = 156 bits (394), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 76/131 (58%), Positives = 100/131 (76%), Gaps = 3/131 (2%)
Query 2 WEQELREKSEE---EKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVN 58
WEQ + + EE E E R++ ++ QTKKDL+PLI L+ + LE +ILEK++ +V
Sbjct 153 WEQHILSQREEFLQEGNVAEARRRDAMLAQTKKDLQPLINMLKSQTLEEEILEKMYNVVI 212
Query 59 LAKERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFI 118
+++ Y+ AH A++LLA+GNA WPMGVTMVGIHERAGRSK+ S+VAHILNDE TRK+I
Sbjct 213 CCEKKDYQAAHEAYMLLAIGNAAWPMGVTMVGIHERAGRSKIFTSEVAHILNDEKTRKYI 272
Query 119 QMLKRLLSFAQ 129
QM+KRLLSFAQ
Sbjct 273 QMIKRLLSFAQ 283
> cpv:cgd7_5490 PRP18 (SFM+PRP18 domains) ; K12817 pre-mRNA-splicing
factor 18
Length=337
Score = 148 bits (373), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 68/139 (48%), Positives = 99/139 (71%), Gaps = 2/139 (1%)
Query 2 WEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELD--ILEKLFVIVNL 59
WE+ L+ + + + T +G Q ++ + QTK+D++PL+ L D ++D +L+KLF IV L
Sbjct 195 WEKLLKCRKKADSETEKGMQDSAQYYQTKRDIQPLVNSLEASDGKVDKEVLDKLFEIVTL 254
Query 60 AKERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQ 119
+R Y +A ++ LA+GNAPWPMGVTMVGIHERAGR+K+ S +AH+LNDETTRK+IQ
Sbjct 255 CNQRDYNKAQDKYIELAIGNAPWPMGVTMVGIHERAGRTKIFSSHIAHVLNDETTRKYIQ 314
Query 120 MLKRLLSFAQRKFPTDPSQ 138
M KRL++ + K P+ Q
Sbjct 315 MFKRLVTHCESKRPSREQQ 333
> mmu:67229 Prpf18, 2810441A10Rik; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 99/140 (70%), Gaps = 0/140 (0%)
Query 1 LWEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLA 60
+W +EL + + K + +G+ ++ +QT+ LRPL ++LR+R+L DI E + I+
Sbjct 198 VWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESITDIIKFM 257
Query 61 KERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQM 120
+R+Y +A+ A++ +A+GNAPWP+GVTMVGIH R GR K+ VAH+LNDET RK+IQ
Sbjct 258 LQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQG 317
Query 121 LKRLLSFAQRKFPTDPSQTV 140
LKRL++ Q+ FPTDPS+ V
Sbjct 318 LKRLMTICQKHFPTDPSKCV 337
> hsa:8559 PRPF18, FLJ10210, PRP18, hPrp18; PRP18 pre-mRNA processing
factor 18 homolog (S. cerevisiae); K12817 pre-mRNA-splicing
factor 18
Length=342
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 66/140 (47%), Positives = 99/140 (70%), Gaps = 0/140 (0%)
Query 1 LWEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLA 60
+W +EL + + K + +G+ ++ +QT+ LRPL ++LR+R+L DI E + I+
Sbjct 198 VWAKELNAREDYVKRSVQGKLNSATQKQTESYLRPLFRKLRKRNLPADIKESITDIIKFM 257
Query 61 KERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQM 120
+R+Y +A+ A++ +A+GNAPWP+GVTMVGIH R GR K+ VAH+LNDET RK+IQ
Sbjct 258 LQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQG 317
Query 121 LKRLLSFAQRKFPTDPSQTV 140
LKRL++ Q+ FPTDPS+ V
Sbjct 318 LKRLMTICQKHFPTDPSKCV 337
> dre:431751 prpf18, zgc:91830; PRP18 pre-mRNA processing factor
18 homolog (yeast); K12817 pre-mRNA-splicing factor 18
Length=342
Score = 143 bits (361), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 98/140 (70%), Gaps = 0/140 (0%)
Query 1 LWEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLA 60
+W ++L + + K + +G+ ++ +QT+ L PL ++LR+++L DI E + I+
Sbjct 198 VWAKDLNSREDHIKRSVQGKLASATQKQTESYLEPLFRKLRKKNLPADIKESITDIIKFM 257
Query 61 KERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQM 120
ER+Y +A+ A++ +A+GNAPWP+GVTMVGIH R GR K+ VAH+LNDET RK+IQ
Sbjct 258 LEREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQG 317
Query 121 LKRLLSFAQRKFPTDPSQTV 140
LKRL++ Q+ FPTDPS+ V
Sbjct 318 LKRLMTICQKHFPTDPSKCV 337
> ath:AT1G03140 splicing factor Prp18 family protein; K12817 pre-mRNA-splicing
factor 18
Length=420
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 66/139 (47%), Positives = 94/139 (67%), Gaps = 0/139 (0%)
Query 2 WEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAK 61
W+QEL E+ T +G+Q + +Q + L PL R++ L DI + L V+VN
Sbjct 240 WKQELDAMENTERRTAKGKQMVATFKQCARYLVPLFNLCRKKGLPADIRQALMVMVNHCI 299
Query 62 ERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQML 121
+R Y A ++ LA+GNAPWP+GVTMVGIHER+ R K+ + VAHI+NDETTRK++Q +
Sbjct 300 KRDYLAAMDHYIKLAIGNAPWPIGVTMVGIHERSAREKIYTNSVAHIMNDETTRKYLQSV 359
Query 122 KRLLSFAQRKFPTDPSQTV 140
KRL++F QR++PT PS+ V
Sbjct 360 KRLMTFCQRRYPTMPSKAV 378
> xla:734335 prpf18, MGC85069; PRP18 pre-mRNA processing factor
18 homolog; K12817 pre-mRNA-splicing factor 18
Length=342
Score = 140 bits (353), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 63/140 (45%), Positives = 97/140 (69%), Gaps = 0/140 (0%)
Query 1 LWEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLA 60
+W +EL + + K + G+ ++ +QT+ L+PL ++LR+++L DI E + I+
Sbjct 198 VWAKELNAREDYVKRSVHGKLASATQKQTESYLKPLFRKLRKKNLPADIKESITDIIKFM 257
Query 61 KERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQM 120
+R+Y +A+ A++ +A+GNAPWP+GVTMVGIH R GR K+ VAH+LNDET RK+IQ
Sbjct 258 LQREYVKANDAYLQMAIGNAPWPIGVTMVGIHARTGREKIFSKHVAHVLNDETQRKYIQG 317
Query 121 LKRLLSFAQRKFPTDPSQTV 140
LKRL++ Q+ F TDPS+ V
Sbjct 318 LKRLMTICQKYFSTDPSKCV 337
> ath:AT1G54590 splicing factor Prp18 family protein
Length=256
Score = 135 bits (341), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 64/140 (45%), Positives = 92/140 (65%), Gaps = 1/140 (0%)
Query 2 WEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAK 61
W+QEL E+ T G+Q + Q + L PL R + L DI + L V+VN
Sbjct 75 WKQELEAMENTERRTAIGKQMLATFNQCARYLTPLFHLCRNKCLPADIRQGLMVMVNCWI 134
Query 62 ERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNV-SQVAHILNDETTRKFIQM 120
+R Y +A F+ LA+GNAPWP+GVTMVGIHER+ R K++ S VAHI+N+ETTRK++Q
Sbjct 135 KRDYLDATAQFIKLAIGNAPWPIGVTMVGIHERSAREKISTSSSVAHIMNNETTRKYLQS 194
Query 121 LKRLLSFAQRKFPTDPSQTV 140
+KRL++F QR++ PS+++
Sbjct 195 VKRLMTFCQRRYSALPSKSI 214
> cel:F32B6.3 hypothetical protein; K12817 pre-mRNA-splicing factor
18
Length=352
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 61/140 (43%), Positives = 91/140 (65%), Gaps = 1/140 (0%)
Query 2 WEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNL-A 60
W ++L ++ + K T +G +A+ H+QT L+ L+ + + + DI L I L
Sbjct 206 WAKDLNDRPLDVKKTAQGMHEAAHHKQTMMHLKSLMTSMERYNCNNDIRHHLAKICRLLV 265
Query 61 KERKYREAHGAFVLLAVGNAPWPMGVTMVGIHERAGRSKLNVSQVAHILNDETTRKFIQM 120
+R Y EA+ A++ +A+GNAPWP+GVT GIH+R G +K VS +AH+LNDET RK+IQ
Sbjct 266 IDRNYLEANNAYMEMAIGNAPWPVGVTRSGIHQRPGSAKSYVSNIAHVLNDETQRKYIQA 325
Query 121 LKRLLSFAQRKFPTDPSQTV 140
KRL++ Q FPTDPS++V
Sbjct 326 FKRLMTKMQEYFPTDPSKSV 345
> sce:YGR006W PRP18; Prp18p; K12817 pre-mRNA-splicing factor 18
Length=251
Score = 61.6 bits (148), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 41/101 (40%), Positives = 64/101 (63%), Gaps = 2/101 (1%)
Query 29 TKKDLRPLIKRLRQRDLELDILEKL-FVIVNLAKERKYREAHGAFVLLAVGNAPWPMGVT 87
TKK L PL+ +LR+ L D+L L V+ +L + ++ A +++ L++GN WP+GVT
Sbjct 139 TKKALFPLLLQLRRNQLAPDLLISLATVLYHLQQPKEINLAVQSYMKLSIGNVAWPIGVT 198
Query 88 MVGIHERAGRSKLNVS-QVAHILNDETTRKFIQMLKRLLSF 127
VGIH R+ SK+ A+I+ DE TR +I +KRL++F
Sbjct 199 SVGIHARSAHSKIQGGRNAANIMIDERTRLWITSIKRLITF 239
> eco:b0727 sucB, ECK0715, JW0716; dihydrolipoyltranssuccinase
(EC:2.3.1.61); K00658 2-oxoglutarate dehydrogenase E2 component
(dihydrolipoamide succinyltransferase) [EC:2.3.1.61]
Length=405
Score = 37.0 bits (84), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 4 QELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRL 40
+E KSEE+ +TP RQQAS+ Q L P I+RL
Sbjct 85 KETSAKSEEKASTPAQRQQASLEEQNNDALSPAIRRL 121
> dre:30660 mtnr1al, mel1a, mel1a2, mtnr1a; melatonin receptor
type 1A like
Length=318
Score = 33.9 bits (76), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 7/65 (10%)
Query 36 LIKRLRQRDLELDILEKLFVIVNLAKERKYREAHGAFVL------LAVGNAPWPMGVTMV 89
L+ + + +D+L L VIV++ + RK R+A AFV+ L V P+P+ +T +
Sbjct 30 LLSSVLITTIVVDVLGNLLVIVSVFRNRKLRKAGNAFVVSLAIADLLVAIYPYPLVLTAI 89
Query 90 GIHER 94
H+R
Sbjct 90 -FHDR 93
> ath:AT5G42870 lipin family protein
Length=930
Score = 31.6 bits (70), Expect = 0.88, Method: Composition-based stats.
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 1 LWEQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLR 41
LW LR ++E +A+P G +Q K RP+ K +R
Sbjct 642 LWPFSLRRSTKEAEASPSGDTAEPEEKQEKSSPRPMKKTVR 682
> mmu:328561 Apol10b, 9130218O11Rik; apolipoprotein L 10b; K14480
apolipoprotein L
Length=334
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 36/124 (29%), Positives = 58/124 (46%), Gaps = 15/124 (12%)
Query 3 EQELREKSEEEKATPEGRQQASIHR----QTKKDLRPLIKRLRQRDLELDILEKLFVIVN 58
++ L ++ +E PE +QQ + KK L I++LR LD + K I N
Sbjct 66 KESLAQEPSDENDGPERKQQNERFLREFPELKKKLEDHIRKLRDLADHLDQVHKDCTISN 125
Query 59 LAKERKYREA-HGAFVLLAVGNAPWPMGVTM------VGIHERAGRSKLNVSQV--AHIL 109
+ + A GA LLA+G P+ GV++ VG+ E A + L + V + L
Sbjct 126 VVS--TFASAISGAMGLLALGLVPFTEGVSLLLSAASVGLGETASVTGLTTTVVEESMRL 183
Query 110 NDET 113
+DE+
Sbjct 184 SDES 187
> hsa:83858 ATAD3B, AAA-TOB3, KIAA1273, TOB3; ATPase family, AAA
domain containing 3B
Length=648
Score = 30.4 bits (67), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 46/152 (30%), Positives = 63/152 (41%), Gaps = 18/152 (11%)
Query 3 EQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAKE 62
E ELR K+E + E R +A R+ +R I RL+ + +LE + L E
Sbjct 177 EMELRHKNEMLRVETEARARAKAERENADIIREQI-RLKASEHRQTVLESIRTAGTLFGE 235
Query 63 ---------RKYREAHGAFVLLAVG--NAPWPMGVTMVGIHERAGRSKLNVSQVAHILND 111
K LLAVG +A VT I R G+ L V + + I
Sbjct 236 GFRAFVTDRDKVTATVAGLTLLAVGVYSAKNATAVTGRFIEARLGKPSL-VRETSRITVL 294
Query 112 ETTRKFIQMLKRLLSFAQRKFPTDPSQTVLLS 143
E R IQ+ +RLLS P D + V+LS
Sbjct 295 EALRHPIQVSRRLLS-----RPQDVLEGVVLS 321
> mmu:75050 Kif27, 4930517I18Rik; kinesin family member 27; K10395
kinesin family member 4/7/21/27
Length=1394
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 1/75 (1%)
Query 30 KKDLRPLIK-RLRQRDLELDILEKLFVIVNLAKERKYREAHGAFVLLAVGNAPWPMGVTM 88
K+DLR L++ +DL + EK ++ AKE + LL VGNA G T
Sbjct 140 KEDLRDLLELETSMKDLHIREDEKGNTVIVGAKECQVESVEDVMSLLQVGNAARHTGTTQ 199
Query 89 VGIHERAGRSKLNVS 103
+ H + +S
Sbjct 200 MNEHSSRSHAIFTIS 214
> xla:734380 atad3a-a, MGC85169, atad3, atad3-a; ATPase family,
AAA domain containing 3A
Length=593
Score = 29.6 bits (65), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 44/152 (28%), Positives = 61/152 (40%), Gaps = 18/152 (11%)
Query 3 EQELREKSEEEKATPEGRQQASIHRQTKKDLRPLIKRLRQRDLELDILEKLFVIVNLAKE 62
E ELR K+E + E R QA + R+ +R I RL+ + +LE + + E
Sbjct 173 EMELRHKNEMLRIEAEARAQAKVERENADIIREQI-RLKAAEHRQTVLESIKTAGTVFGE 231
Query 63 ---------RKYREAHGAFVLLAVG--NAPWPMGVTMVGIHERAGRSKLNVSQVAHILND 111
K LLAVG A GV I R G+ L V + I
Sbjct 232 GFRTFISDWDKVTATVAGLTLLAVGVYTAKNATGVAGRYIEARLGKPSL-VRDTSRITVA 290
Query 112 ETTRKFIQMLKRLLSFAQRKFPTDPSQTVLLS 143
E + I++ KRL S Q D + V+LS
Sbjct 291 EAVKHPIKITKRLYSKIQ-----DALEGVILS 317
> hsa:55582 KIF27, DKFZp434D0917, RP11-575L7.3; kinesin family
member 27; K10395 kinesin family member 4/7/21/27
Length=1401
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/75 (29%), Positives = 33/75 (44%), Gaps = 1/75 (1%)
Query 30 KKDLRPLIK-RLRQRDLELDILEKLFVIVNLAKERKYREAHGAFVLLAVGNAPWPMGVTM 88
K+DLR L++ +DL + EK ++ AKE A LL +GNA G T
Sbjct 140 KEDLRDLLELETSMKDLHIREDEKGNTVIVGAKECHVESAGEVMSLLEMGNAARHTGTTQ 199
Query 89 VGIHERAGRSKLNVS 103
+ H + +S
Sbjct 200 MNEHSSRSHAIFTIS 214
Lambda K H
0.320 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2938175820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40