bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2081_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
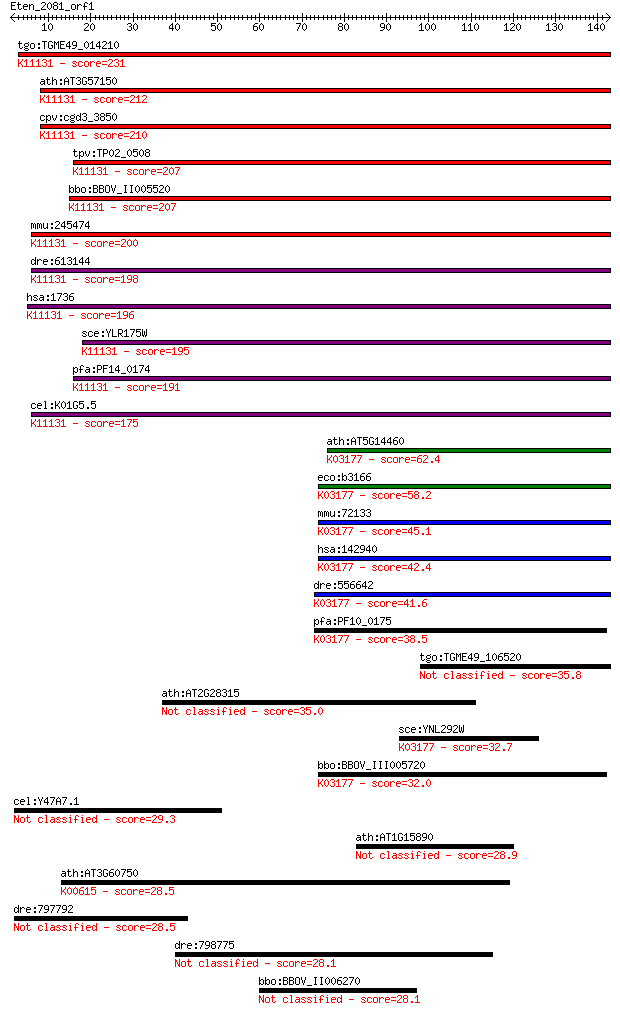
tgo:TGME49_014210 centromere/microtubule binding protein, puta... 231 5e-61
ath:AT3G57150 NAP57; NAP57 (Arabidopsis thaliana homologue of ... 212 3e-55
cpv:cgd3_3850 Cbf5p; centromere-binding factor 5 like PUA doma... 210 1e-54
tpv:TP02_0508 rRNA pseudouridine synthase; K11131 H/ACA ribonu... 207 1e-53
bbo:BBOV_II005520 18.m06459; rRNA pseudouridine synthase famil... 207 1e-53
mmu:245474 Dkc1, BC068171, MGC107501; dyskeratosis congenita 1... 200 2e-51
dre:613144 dkc1, MGC110395, fv62a07, wu:fa28f10, wu:fc87a02, w... 198 5e-51
hsa:1736 DKC1, CBF5, DKC, FLJ97620, NAP57, NOLA4, XAP101; dysk... 196 2e-50
sce:YLR175W CBF5; Cbf5p (EC:5.4.99.-); K11131 H/ACA ribonucleo... 195 3e-50
pfa:PF14_0174 pseudouridine synthase, putative; K11131 H/ACA r... 191 8e-49
cel:K01G5.5 hypothetical protein; K11131 H/ACA ribonucleoprote... 175 4e-44
ath:AT5G14460 pseudouridine synthase/ transporter; K03177 tRNA... 62.4 4e-10
eco:b3166 truB, ECK3155, JW3135, yhbA; tRNA U55 pseudouridine ... 58.2 8e-09
mmu:72133 Trub1, 2610009I02Rik, 9030425C13Rik, AI448235, BB118... 45.1 7e-05
hsa:142940 TRUB1, PUS4; TruB pseudouridine (psi) synthase homo... 42.4 4e-04
dre:556642 trub1, zgc:195155; TruB pseudouridine (psi) synthas... 41.6 8e-04
pfa:PF10_0175 tRNA pseudouridine synthase, putative; K03177 tR... 38.5 0.007
tgo:TGME49_106520 tRNA pseudouridine synthase B, putative (EC:... 35.8 0.048
ath:AT2G28315 hypothetical protein 35.0 0.081
sce:YNL292W PUS4; Pseudouridine synthase, catalyzes only the f... 32.7 0.35
bbo:BBOV_III005720 17.m07509; tRNA pseudouridine synthase; K03... 32.0 0.74
cel:Y47A7.1 hypothetical protein 29.3 3.8
ath:AT1G15890 disease resistance protein (CC-NBS-LRR class), p... 28.9 5.4
ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 tra... 28.5 6.3
dre:797792 si:dkey-259k14.2 28.5 7.6
dre:798775 novel coagulation factor 28.1 8.5
bbo:BBOV_II006270 18.m10022; hypothetical protein 28.1 9.8
> tgo:TGME49_014210 centromere/microtubule binding protein, putative
; K11131 H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=505
Score = 231 bits (589), Expect = 5e-61, Method: Compositional matrix adjust.
Identities = 110/140 (78%), Positives = 123/140 (87%), Gaps = 0/140 (0%)
Query 3 VDTMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSP 62
V + G + KK K + + I+P S++PPIDTSKWPLLLK+YDRLNVRTGHYTPLP G SP
Sbjct 11 VASAGTEGGKKKKKTDNYAIEPNSSSPPIDTSKWPLLLKHYDRLNVRTGHYTPLPHGCSP 70
Query 63 LARPLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINR 122
L RPLQQ++NYG++ LDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINR
Sbjct 71 LCRPLQQYINYGAMNLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINR 130
Query 123 ATRLVKSQQSAGKEYVAVAR 142
ATRLVKSQQSAGKEYV + R
Sbjct 131 ATRLVKSQQSAGKEYVCIIR 150
> ath:AT3G57150 NAP57; NAP57 (Arabidopsis thaliana homologue of
NAP57); pseudouridine synthase; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=565
Score = 212 bits (539), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 98/135 (72%), Positives = 114/135 (84%), Gaps = 0/135 (0%)
Query 8 EKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPL 67
+K + AA ++I+P S P IDTS+WP+LLKNYDRLNVRTGHYTP+ G SPL RPL
Sbjct 15 DKTENDAADTGDYMIKPQSFTPAIDTSQWPILLKNYDRLNVRTGHYTPISAGHSPLKRPL 74
Query 68 QQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLV 127
Q+++ YG + LDKPANPSSHEVVAW+K+ILRVEKTGHSGTLDPKVTG L+VCI+RATRLV
Sbjct 75 QEYIRYGVINLDKPANPSSHEVVAWIKRILRVEKTGHSGTLDPKVTGNLIVCIDRATRLV 134
Query 128 KSQQSAGKEYVAVAR 142
KSQQ AGKEYV VAR
Sbjct 135 KSQQGAGKEYVCVAR 149
> cpv:cgd3_3850 Cbf5p; centromere-binding factor 5 like PUA domain
containing protein with a type I pseudouridine synthase
domain ; K11131 H/ACA ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=520
Score = 210 bits (534), Expect = 1e-54, Method: Compositional matrix adjust.
Identities = 101/135 (74%), Positives = 113/135 (83%), Gaps = 1/135 (0%)
Query 8 EKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPL 67
E E+ K E VI+P S + PIDTS WPLLLKNY +LNVRTGHYTPLP+G+SPL R L
Sbjct 8 ELEEMSDGKIE-TVIKPESKSQPIDTSNWPLLLKNYSKLNVRTGHYTPLPYGNSPLNRNL 66
Query 68 QQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLV 127
+L+YG + LDKPANPSSHEVVAW+KKILR EKTGHSGTLDPKVTGCLLVC+NRATRLV
Sbjct 67 ADYLSYGIINLDKPANPSSHEVVAWIKKILRCEKTGHSGTLDPKVTGCLLVCLNRATRLV 126
Query 128 KSQQSAGKEYVAVAR 142
KSQQSAGKEYV + R
Sbjct 127 KSQQSAGKEYVCIVR 141
> tpv:TP02_0508 rRNA pseudouridine synthase; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=421
Score = 207 bits (527), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 93/127 (73%), Positives = 111/127 (87%), Gaps = 0/127 (0%)
Query 16 KAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGS 75
K + F+I+P + +PPIDTS WPLLLKNYD+LNVRTGHYTP+P GSSPL R ++ ++ YG
Sbjct 3 KNKSFLIKPEANSPPIDTSNWPLLLKNYDKLNVRTGHYTPMPHGSSPLCRDMKNYMKYGY 62
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGK 135
+ LDKPANPSSHEVVAW+K+ILR EKTGHSGTLDPKVTG LLVC+NR+TRLVKSQQ+ GK
Sbjct 63 INLDKPANPSSHEVVAWIKRILRCEKTGHSGTLDPKVTGVLLVCLNRSTRLVKSQQTHGK 122
Query 136 EYVAVAR 142
EYVAV +
Sbjct 123 EYVAVVK 129
> bbo:BBOV_II005520 18.m06459; rRNA pseudouridine synthase family
protein; K11131 H/ACA ribonucleoprotein complex subunit
4 [EC:5.4.99.-]
Length=418
Score = 207 bits (526), Expect = 1e-53, Method: Compositional matrix adjust.
Identities = 96/128 (75%), Positives = 110/128 (85%), Gaps = 0/128 (0%)
Query 15 AKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYG 74
AK FVI+P APPIDTS+WPLLLKNYD+LNVRTGHYTPLP GSSPL R L+ ++ YG
Sbjct 2 AKDGDFVIKPQKAAPPIDTSEWPLLLKNYDKLNVRTGHYTPLPHGSSPLCRDLRNYIKYG 61
Query 75 SLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAG 134
+ LDKPANPSSHE V+W+K+ILR EKTGHSGTLDPKVTG LLVCI+RATRLVKSQQ+ G
Sbjct 62 FINLDKPANPSSHETVSWIKRILRCEKTGHSGTLDPKVTGVLLVCIDRATRLVKSQQTHG 121
Query 135 KEYVAVAR 142
KEYVA+ +
Sbjct 122 KEYVAIVK 129
> mmu:245474 Dkc1, BC068171, MGC107501; dyskeratosis congenita
1, dyskerin homolog (human); K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=509
Score = 200 bits (508), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 91/137 (66%), Positives = 111/137 (81%), Gaps = 0/137 (0%)
Query 6 MGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLAR 65
+ E + + AE F+I+P S +DTS+WPLLLKN+D+LNVRT HYTPLP GS+PL R
Sbjct 22 LQEDDVAEIQHAEEFLIKPESKVAQLDTSQWPLLLKNFDKLNVRTAHYTPLPCGSNPLKR 81
Query 66 PLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATR 125
+ ++ G + LDKP+NPSSHEVVAW+++ILRVEKTGHSGTLDPKVTGCL+VCI RATR
Sbjct 82 EIGDYIRTGFINLDKPSNPSSHEVVAWIRRILRVEKTGHSGTLDPKVTGCLIVCIERATR 141
Query 126 LVKSQQSAGKEYVAVAR 142
LVKSQQSAGKEYV + R
Sbjct 142 LVKSQQSAGKEYVGIVR 158
> dre:613144 dkc1, MGC110395, fv62a07, wu:fa28f10, wu:fc87a02,
wu:fi24a05, wu:fv62a07, zgc:110395; dyskeratosis congenita
1, dyskerin; K11131 H/ACA ribonucleoprotein complex subunit
4 [EC:5.4.99.-]
Length=506
Score = 198 bits (503), Expect = 5e-51, Method: Compositional matrix adjust.
Identities = 89/137 (64%), Positives = 111/137 (81%), Gaps = 0/137 (0%)
Query 6 MGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLAR 65
+ ++E ++ F+I+P S +DTS+WPLLLKN+D+LN+RT HYTPLP GS+PL R
Sbjct 17 ISDEEVGDIQESGDFLIKPESKVASLDTSQWPLLLKNFDKLNIRTAHYTPLPHGSNPLKR 76
Query 66 PLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATR 125
+ ++ G + LDKPANPSSHEVVAW+K+ILRVEKTGHSGTLDPKVTGCL+VC+ RATR
Sbjct 77 GIHDYVRSGFINLDKPANPSSHEVVAWIKRILRVEKTGHSGTLDPKVTGCLIVCVERATR 136
Query 126 LVKSQQSAGKEYVAVAR 142
LVKSQQSAGKEYV + R
Sbjct 137 LVKSQQSAGKEYVGIVR 153
> hsa:1736 DKC1, CBF5, DKC, FLJ97620, NAP57, NOLA4, XAP101; dyskeratosis
congenita 1, dyskerin; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=509
Score = 196 bits (499), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 90/138 (65%), Positives = 112/138 (81%), Gaps = 0/138 (0%)
Query 5 TMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLA 64
++ E++ + AE F+I+P S +DTS+WPLLLKN+D+LNVRT HYTPL GS+PL
Sbjct 21 SLPEEDVAEIQHAEEFLIKPESKVAKLDTSQWPLLLKNFDKLNVRTTHYTPLACGSNPLK 80
Query 65 RPLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRAT 124
R + ++ G + LDKP+NPSSHEVVAW+++ILRVEKTGHSGTLDPKVTGCL+VCI RAT
Sbjct 81 REIGDYIRTGFINLDKPSNPSSHEVVAWIRRILRVEKTGHSGTLDPKVTGCLIVCIERAT 140
Query 125 RLVKSQQSAGKEYVAVAR 142
RLVKSQQSAGKEYV + R
Sbjct 141 RLVKSQQSAGKEYVGIVR 158
> sce:YLR175W CBF5; Cbf5p (EC:5.4.99.-); K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=483
Score = 195 bits (496), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 88/125 (70%), Positives = 106/125 (84%), Gaps = 0/125 (0%)
Query 18 EGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGSLL 77
E FVI+P + DTS+WPLLLKN+D+L VR+GHYTP+P GSSPL R L+ +++ G +
Sbjct 4 EDFVIKPEAAGASTDTSEWPLLLKNFDKLLVRSGHYTPIPAGSSPLKRDLKSYISSGVIN 63
Query 78 LDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEY 137
LDKP+NPSSHEVVAW+K+ILR EKTGHSGTLDPKVTGCL+VCI+RATRLVKSQQ AGKEY
Sbjct 64 LDKPSNPSSHEVVAWIKRILRCEKTGHSGTLDPKVTGCLIVCIDRATRLVKSQQGAGKEY 123
Query 138 VAVAR 142
V + R
Sbjct 124 VCIVR 128
> pfa:PF14_0174 pseudouridine synthase, putative; K11131 H/ACA
ribonucleoprotein complex subunit 4 [EC:5.4.99.-]
Length=466
Score = 191 bits (484), Expect = 8e-49, Method: Compositional matrix adjust.
Identities = 85/127 (66%), Positives = 104/127 (81%), Gaps = 0/127 (0%)
Query 16 KAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGS 75
K + I+P IDTS WPLLLKNYD+LN+RT H+TPLP G+SP +R ++++L YG
Sbjct 4 KKMDYKIEPERKEAAIDTSNWPLLLKNYDKLNIRTSHFTPLPMGNSPYSRNIEEYLKYGI 63
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGK 135
+ LDKP+NPSSHEVV+W++KILR EKTGHSGTLDPKVTG LLVC+NRATRLVKSQQ +GK
Sbjct 64 INLDKPSNPSSHEVVSWIRKILRCEKTGHSGTLDPKVTGVLLVCLNRATRLVKSQQESGK 123
Query 136 EYVAVAR 142
EYV V +
Sbjct 124 EYVCVCK 130
> cel:K01G5.5 hypothetical protein; K11131 H/ACA ribonucleoprotein
complex subunit 4 [EC:5.4.99.-]
Length=445
Score = 175 bits (443), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 81/146 (55%), Positives = 107/146 (73%), Gaps = 9/146 (6%)
Query 6 MGEKEKK---------KAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPL 56
MG+K+K+ +A + F + ++ +D S+WPLLLKNYD+LNVRT HYTP
Sbjct 1 MGKKDKRSKLEGDDLAEAQQKGSFQLPSSNETAKLDASQWPLLLKNYDKLNVRTNHYTPH 60
Query 57 PFGSSPLARPLQQHLNYGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCL 116
G SPL R ++ +++ G LDKP+NPSSHEVV+W+K+ILR EKTGHSGTLDPKV+GCL
Sbjct 61 VEGVSPLKRDIKNYISSGFFNLDKPSNPSSHEVVSWIKRILRCEKTGHSGTLDPKVSGCL 120
Query 117 LVCINRATRLVKSQQSAGKEYVAVAR 142
+VCI+R TRL KSQQ AGKEY+ + +
Sbjct 121 IVCIDRTTRLAKSQQGAGKEYICIFK 146
> ath:AT5G14460 pseudouridine synthase/ transporter; K03177 tRNA
pseudouridine synthase B [EC:5.4.99.12]
Length=540
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 44/67 (65%), Gaps = 0/67 (0%)
Query 76 LLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGK 135
LL++KP +S V L+++++V+K GH+GTLDP TG L+VC+ +AT++V Q K
Sbjct 324 LLVNKPKGWTSFTVCGKLRRLVKVKKVGHAGTLDPMATGLLIVCVGKATKVVDRYQGMIK 383
Query 136 EYVAVAR 142
Y V R
Sbjct 384 GYSGVFR 390
> eco:b3166 truB, ECK3155, JW3135, yhbA; tRNA U55 pseudouridine
synthase (EC:4.2.1.70); K03177 tRNA pseudouridine synthase
B [EC:5.4.99.12]
Length=314
Score = 58.2 bits (139), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 27/69 (39%), Positives = 40/69 (57%), Gaps = 0/69 (0%)
Query 74 GSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSA 133
G LLLDKP SS++ + +K+I + GH+G LDP TG L +C+ AT+ + +
Sbjct 13 GVLLLDKPQGMSSNDALQKVKRIYNANRAGHTGALDPLATGMLPICLGEATKFSQYLLDS 72
Query 134 GKEYVAVAR 142
K Y +AR
Sbjct 73 DKRYRVIAR 81
> mmu:72133 Trub1, 2610009I02Rik, 9030425C13Rik, AI448235, BB118943,
C76870, MGC107612; TruB pseudouridine (psi) synthase
homolog 1 (E. coli); K03177 tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=338
Score = 45.1 bits (105), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 38/85 (44%), Gaps = 16/85 (18%)
Query 74 GSLLLDKPANPSSHEVVAWLKKILRVE----------------KTGHSGTLDPKVTGCLL 117
G + KP P+S E++ LK+ L E K GH GTLD G L+
Sbjct 58 GVFAVHKPKGPTSAELLNRLKEKLLAEAGMPSPEWNKRQKQTLKVGHGGTLDSAAQGVLV 117
Query 118 VCINRATRLVKSQQSAGKEYVAVAR 142
V I R T+++ S S K Y+ +
Sbjct 118 VGIGRGTKMLTSMLSGSKRYITIGE 142
> hsa:142940 TRUB1, PUS4; TruB pseudouridine (psi) synthase homolog
1 (E. coli); K03177 tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=349
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 27/85 (31%), Positives = 37/85 (43%), Gaps = 16/85 (18%)
Query 74 GSLLLDKPANPSSHEVVAWLKKILRVE----------------KTGHSGTLDPKVTGCLL 117
G + KP P+S E++ LK+ L E K GH GTLD G L+
Sbjct 70 GVFAVHKPKGPTSAELLNRLKEKLLAEAGMPSPEWTKRKKQTLKIGHGGTLDSAARGVLV 129
Query 118 VCINRATRLVKSQQSAGKEYVAVAR 142
V I T+++ S S K Y A+
Sbjct 130 VGIGSGTKMLTSMLSGSKRYTAIGE 154
> dre:556642 trub1, zgc:195155; TruB pseudouridine (psi) synthase
homolog 1 (E. coli); K03177 tRNA pseudouridine synthase
B [EC:5.4.99.12]
Length=290
Score = 41.6 bits (96), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 38/85 (44%), Gaps = 15/85 (17%)
Query 73 YGSLLLDKPANPSSHEVVAWLKKILRVE---------------KTGHSGTLDPKVTGCLL 117
+G + K P+S +V+ LKK L E K GH GTLD +G L+
Sbjct 19 HGIFAIHKKKGPTSADVLNILKKTLLKEAGVTNFNPRKRKQPFKIGHGGTLDSNASGVLV 78
Query 118 VCINRATRLVKSQQSAGKEYVAVAR 142
V I T+++ + + K+Y A
Sbjct 79 VGIGEGTKMLTTMLAGSKKYTAFGE 103
> pfa:PF10_0175 tRNA pseudouridine synthase, putative; K03177
tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=560
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 27/81 (33%), Positives = 39/81 (48%), Gaps = 12/81 (14%)
Query 73 YGSLL-LDKPANPSSHEVVAWLKKILRVE-----------KTGHSGTLDPKVTGCLLVCI 120
YG L + KP S V +KKIL+ K GH GTLDP G L++ I
Sbjct 325 YGGFLNIYKPVKLYSMRVCEKIKKILKDYFYKLNKKKVNIKVGHGGTLDPFAEGVLIIGI 384
Query 121 NRATRLVKSQQSAGKEYVAVA 141
+AT+ + K+Y+A++
Sbjct 385 QQATKKLSDFLKCYKKYLALS 405
> tgo:TGME49_106520 tRNA pseudouridine synthase B, putative (EC:3.2.1.8)
Length=573
Score = 35.8 bits (81), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 98 RVEKTGHSGTLDPKVTGCLLVCINRATRLVKSQQSAGKEYVAVAR 142
R GH GTLDP TG L++ I T+ ++ + K Y AV R
Sbjct 318 RKTGVGHGGTLDPAATGVLILGIGEGTKRLRHFLTGWKRYKAVVR 362
> ath:AT2G28315 hypothetical protein
Length=342
Score = 35.0 bits (79), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 21/74 (28%), Positives = 38/74 (51%), Gaps = 1/74 (1%)
Query 37 PLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNYGSLLLDKPANPSSHEVVAWLKKI 96
P + K LNV + HY+P+ G L+ + +N+ + L+ +P +++V+ LK
Sbjct 202 PFVDKYLTSLNVFSFHYSPIVVGFITLSCLIAVSVNFSTFLVIGKTSPVTYQVLGHLKTC 261
Query 97 LRVEKTGHSGTLDP 110
L V G++ DP
Sbjct 262 L-VLAFGYTLLHDP 274
> sce:YNL292W PUS4; Pseudouridine synthase, catalyzes only the
formation of pseudouridine-55 (Psi55), a highly conserved tRNA
modification, in mitochondrial and cytoplasmic tRNAs; PUS4
overexpression leads to translational derepression of GCN4
(Gcd-phenotype) (EC:4.2.1.70); K03177 tRNA pseudouridine synthase
B [EC:5.4.99.12]
Length=403
Score = 32.7 bits (73), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 1/33 (3%)
Query 93 LKKILRVEKTGHSGTLDPKVTGCLLVCINRATR 125
L+K+ +V K GH GTLDP +G L++ I T+
Sbjct 60 LRKVSKV-KMGHGGTLDPLASGVLVIGIGAGTK 91
> bbo:BBOV_III005720 17.m07509; tRNA pseudouridine synthase; K03177
tRNA pseudouridine synthase B [EC:5.4.99.12]
Length=282
Score = 32.0 bits (71), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 13/81 (16%)
Query 74 GSLLLDKPANPSSHEVVAWLKKILRVE-------------KTGHSGTLDPKVTGCLLVCI 120
G L + KP +S++ +KKI+ E K GH GTLD G L + I
Sbjct 34 GILNIYKPVMITSNDACQMVKKIIIDEMFKRKTIINKPKIKVGHGGTLDMHAEGVLAIGI 93
Query 121 NRATRLVKSQQSAGKEYVAVA 141
T+++ K Y A+
Sbjct 94 GNGTKMLPYYLKGDKIYEAIG 114
> cel:Y47A7.1 hypothetical protein
Length=1114
Score = 29.3 bits (64), Expect = 3.8, Method: Composition-based stats.
Identities = 14/49 (28%), Positives = 27/49 (55%), Gaps = 8/49 (16%)
Query 2 YVDTMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRT 50
++ GE++K++AA+A + ++ WP +L+N+D LN T
Sbjct 98 FMQNQGEEKKEQAAQARRRI--------SVELKLWPEILENFDVLNFNT 138
> ath:AT1G15890 disease resistance protein (CC-NBS-LRR class),
putative
Length=851
Score = 28.9 bits (63), Expect = 5.4, Method: Composition-based stats.
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 0/37 (0%)
Query 83 NPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLVC 119
N SSHE + +KIL V K + D KV C L C
Sbjct 376 NSSSHEFPSMEEKILPVLKFSYDDLKDEKVKLCFLYC 412
> ath:AT3G60750 transketolase, putative (EC:2.2.1.1); K00615 transketolase
[EC:2.2.1.1]
Length=741
Score = 28.5 bits (62), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 22/106 (20%)
Query 13 KAAKAEGFVIQPTSTAPPIDTSKWPLLLKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLN 72
+AA E ++PT+ + +D S + D + + LP G +P+A H+
Sbjct 65 RAAAVE--TVEPTTDSSIVDKSVNSIRFLAIDAVEKAKSGHPGLPMGCAPMA-----HIL 117
Query 73 YGSLLLDKPANPSSHEVVAWLKKILRVEKTGHSGTLDPKVTGCLLV 118
Y ++ P NP W + V GH GC+L+
Sbjct 118 YDEVMRYNPKNPY------WFNRDRFVLSAGH---------GCMLL 148
> dre:797792 si:dkey-259k14.2
Length=609
Score = 28.5 bits (62), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 22/44 (50%), Gaps = 3/44 (6%)
Query 2 YVDTMGEKEKKKAAKAEGFVIQPTSTAPPIDTSKWPL---LLKN 42
Y DT+ +EK A KA+G V + P + KW LLKN
Sbjct 325 YFDTVTSEEKDPACKADGLVTFERVSVPASELPKWKSEKKLLKN 368
> dre:798775 novel coagulation factor
Length=427
Score = 28.1 bits (61), Expect = 8.5, Method: Composition-based stats.
Identities = 21/87 (24%), Positives = 37/87 (42%), Gaps = 12/87 (13%)
Query 40 LKNYDRLNVRTGHYTPLPFGSSPLARPLQQHLNY------------GSLLLDKPANPSSH 87
L+ + +VR G Y F S + P++QH+++ L L+ PA S++
Sbjct 231 LETSSKFSVRLGDYQRFRFEGSEITLPVKQHISHPQYNPITVDNDIALLRLEVPAKFSTY 290
Query 88 EVVAWLKKILRVEKTGHSGTLDPKVTG 114
+ A L + E+ H +TG
Sbjct 291 ILPACLPSLELAERMLHRNGTVTVITG 317
> bbo:BBOV_II006270 18.m10022; hypothetical protein
Length=1171
Score = 28.1 bits (61), Expect = 9.8, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 20/42 (47%), Gaps = 5/42 (11%)
Query 60 SSPLARPLQQHLNYG-----SLLLDKPANPSSHEVVAWLKKI 96
S PLA P +H+ + LLL P +PS V WL I
Sbjct 601 SHPLAEPSTEHIRHNCVAGFELLLRTPWSPSIDSVFLWLSMI 642
Lambda K H
0.315 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2683748972
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40