bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2079_orf1
Length=209
Score E
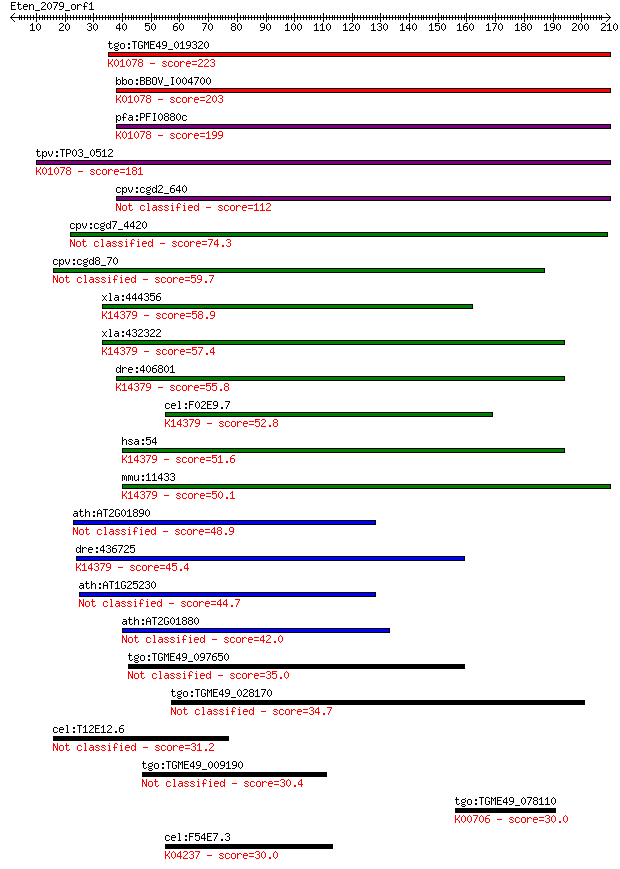
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01... 223 4e-58
bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01... 203 3e-52
pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3... 199 8e-51
tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosp... 181 2e-45
cpv:cgd2_640 acid phosphatase 112 6e-25
cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),s... 74.3 3e-13
cpv:cgd8_70 hypothetical protein 59.7 8e-09
xla:444356 MGC82831 protein; K14379 tartrate-resistant acid ph... 58.9 1e-08
xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resist... 57.4 4e-08
dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:f... 55.8 1e-07
cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant ac... 52.8 9e-07
hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate res... 51.6 2e-06
mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resi... 50.1 5e-06
ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid pho... 48.9 1e-05
dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate res... 45.4 1e-04
ath:AT1G25230 purple acid phosphatase family protein 44.7 2e-04
ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid pho... 42.0 0.002
tgo:TGME49_097650 serine/threonine protein phosphatase, putati... 35.0 0.19
tgo:TGME49_028170 serine/threonine protein phosphatase, putati... 34.7 0.24
cel:T12E12.6 hypothetical protein 31.2 2.9
tgo:TGME49_009190 ABC transporter, putative (EC:3.6.3.44) 30.4 4.4
tgo:TGME49_078110 1,3-beta-glucan synthase component domain-co... 30.0 5.8
cel:F54E7.3 par-3; abnormal embryonic PARtitioning of cytoplas... 30.0 6.5
> tgo:TGME49_019320 acid phosphatase, putative (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=431
Score = 223 bits (568), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 110/191 (57%), Positives = 133/191 (69%), Gaps = 16/191 (8%)
Query 35 ASLAQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSNFPNGVSGANDTKWD 94
A +AQL+F LGN+G GQK VA+TLKK AA E ISF+ SPGSNF GVS NDT+W
Sbjct 47 AVVAQLKFVGLGNWGSGSYGQKTVADTLKKVAANEHISFIASPGSNFLGGVSSLNDTRWQ 106
Query 95 EMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYG-------DGSSEEAVSN 147
FE+VYSD G+L+MPF TVLG +DWS NYT+ RT+L Y DG A +
Sbjct 107 SEFENVYSDANGALKMPFFTVLGVDDWSRNYTSEALRTELTYAVTSEQIKDGKLAPADAT 166
Query 148 S---------PKWTLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLSSAFPF 198
PKWTLPNWWYHY +HF A+TG AF++SGHKDMSVG++F+DTWVLSS+FPF
Sbjct 167 EAAAAENHGYPKWTLPNWWYHYLMHFPANTGGAFINSGHKDMSVGMIFIDTWVLSSSFPF 226
Query 199 GSVTEKAWKDL 209
+VT +AW DL
Sbjct 227 SNVTSRAWADL 237
> bbo:BBOV_I004700 19.m02068; acid phosphatase (EC:3.1.3.2); K01078
acid phosphatase [EC:3.1.3.2]
Length=395
Score = 203 bits (516), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 91/174 (52%), Positives = 118/174 (67%), Gaps = 2/174 (1%)
Query 38 AQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSNFPNGVSGANDTKWDEMF 97
AQLRFA++GN+G QK VAETLKK+ A +R++F+VSPGSNF GV+G+NDTKWD F
Sbjct 27 AQLRFASVGNWGTGSKYQKRVAETLKKSIANDRVTFIVSPGSNFEYGVTGSNDTKWDTHF 86
Query 98 EDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEE--AVSNSPKWTLPN 155
+ VY E GS+ +P TVLGA DW G++ + R Y +E P+WT+PN
Sbjct 87 QSVYRSEDGSMEIPMFTVLGAGDWLGDFNSQINRNQQAYFTSQVDEKNGAKGLPRWTMPN 146
Query 156 WWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLSSAFPFGSVTEKAWKDL 209
WWYHY HF + + L SGHKDMSVG +F+DTW+LS+AFP+ V+ AW DL
Sbjct 147 WWYHYYTHFATTASMSLLKSGHKDMSVGFIFIDTWILSTAFPYKDVSNAAWADL 200
> pfa:PFI0880c GAP50; glideosome-associated protein 50 (EC:3.1.3.2);
K01078 acid phosphatase [EC:3.1.3.2]
Length=396
Score = 199 bits (505), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 87/178 (48%), Positives = 120/178 (67%), Gaps = 6/178 (3%)
Query 38 AQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSNFPNGVSGANDTKWDEMF 97
QLRFA+LG++G D GQ A+ K+ ER++F+VSPGSNF +GV G ND W ++
Sbjct 25 CQLRFASLGDWGKDTKGQILNAKYFKQFIKNERVTFIVSPGSNFIDGVKGLNDPAWKNLY 84
Query 98 EDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAV------SNSPKW 151
EDVYS+EKG + MPF TVLG DW+GNY A + +Y + + E ++ +N PKW
Sbjct 85 EDVYSEEKGDMYMPFFTVLGTRDWTGNYNAQLLKGQGIYIEKNGETSIEKDADATNYPKW 144
Query 152 TLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLSSAFPFGSVTEKAWKDL 209
+PN+WYHY HFT S+G + + +GHKD++ +F+DTWVLSS FP+ + EKAW DL
Sbjct 145 IMPNYWYHYFTHFTVSSGPSIVKTGHKDLAAAFIFIDTWVLSSNFPYKKIHEKAWNDL 202
> tpv:TP03_0512 acid phosphatase (EC:3.1.3.2); K01078 acid phosphatase
[EC:3.1.3.2]
Length=404
Score = 181 bits (459), Expect = 2e-45, Method: Compositional matrix adjust.
Identities = 89/212 (41%), Positives = 126/212 (59%), Gaps = 12/212 (5%)
Query 10 LRFASCRVRPVALAAAILVGLLEIPASLAQLRFAALGNYGFDISGQKEVAETLKKTAAEE 69
++ +R L + L+ + + A LRFA+LGN+G QK VAE LK+ E
Sbjct 1 MKLKIISIRYFMLYSLPLLIFIFSSSVNASLRFASLGNWGTGSKTQKLVAEKLKEYVKNE 60
Query 70 RISFLVSPGSNFPNGVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMR 129
R+++L+SPG NF NGV+G ND KW + FE VY+D+ G + +P TVLG+EDW G+Y A
Sbjct 61 RVTYLLSPGFNFDNGVNGLNDEKWKKFFESVYNDDSGLMDLPMFTVLGSEDWLGDYNAQY 120
Query 130 ERTDLVYGDG------------SSEEAVSNSPKWTLPNWWYHYAVHFTASTGSAFLSSGH 177
ER Y +G S S++P+ +PNWWYH+ F+ + + L SGH
Sbjct 121 ERYHQFYLNGHLPKDLSSVDYKSDSNNSSHNPRLIMPNWWYHFFTTFSTNASVSLLKSGH 180
Query 178 KDMSVGLVFVDTWVLSSAFPFGSVTEKAWKDL 209
KD+SV VFVDTWVLS+ FP+ V+ +AW +L
Sbjct 181 KDLSVAFVFVDTWVLSNQFPYKDVSNEAWNEL 212
> cpv:cgd2_640 acid phosphatase
Length=339
Score = 112 bits (281), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 65/173 (37%), Positives = 104/173 (60%), Gaps = 19/173 (10%)
Query 38 AQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSNFPNGVSGANDTKWDEMF 97
+L FA+L NYG QK+VA LK A + S LVSPG NFP G+ + F
Sbjct 23 GELYFASLSNYGCS-GNQKKVASVLKAQAEKTPFSLLVSPGDNFPGGID------FKHCF 75
Query 98 EDVYSDEKGSLRMPFLTVLGAEDW-SGNYTAMRERTDLVYGDGSSEEAVSNSPKWTLPNW 156
E++YS++ SL++P +G DW +GN + +R ++ Y D +++ P+++ PN+
Sbjct 76 ENIYSEK--SLQIPLFAAMGQADWDNGNANLLLKRNNVTY-DSNNDIF----PRFSFPNY 128
Query 157 WYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLSSAFPFGSVTEKAWKDL 209
+YHY H+T + S LS +D +V VF+DT++LSS+FP V+E+A+++L
Sbjct 129 FYHYVSHYTDT--SNVLS--RRDGTVLFVFIDTFILSSSFPDHKVSEQAFQNL 177
> cpv:cgd7_4420 secreted acid phosphatase (calcineurin family),signal
peptide
Length=826
Score = 74.3 bits (181), Expect = 3e-13, Method: Composition-based stats.
Identities = 58/201 (28%), Positives = 95/201 (47%), Gaps = 28/201 (13%)
Query 22 LAAAILVGLLEIP-ASLAQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSN 80
L A L+ LL + S L + G++G + V+ ++ A+ + +F++S G N
Sbjct 8 LLRATLICLLAVTRCSSEPLSWMTFGDWGEPTAILSAVSRSMANLASIIKPNFIISVGDN 67
Query 81 FPN-GVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDG 139
F GVS +D W+ MFE V+ D++ + F VLG DW GN TA +R
Sbjct 68 FYRWGVSSVDDPIWENMFESVF-DQESLQDVQFRCVLGNHDWWGNATAQVDR-------- 118
Query 140 SSEEAVSNSPKWTLPNWWYHYAVHFTASTGSAF----LSSGHKDMSVGLV-------FVD 188
SP+W LPN+WY+ F + S +SS + + +V + D
Sbjct 119 ---HYSLKSPRWYLPNFWYYTIEEFESPVNSPHPYLNVSSSPTEETEEMVKTKAIFIYTD 175
Query 189 TWVLSSAFPFGS-VTEKAWKD 208
+W++SS P G+ +T + W +
Sbjct 176 SWIISS--PMGTDITPELWNE 194
> cpv:cgd8_70 hypothetical protein
Length=424
Score = 59.7 bits (143), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 48/174 (27%), Positives = 77/174 (44%), Gaps = 18/174 (10%)
Query 16 RVRPVALAAAILVGLLEIPASLAQLRFAALGNYGFDISGQKEVAETLKKTAAEERISFLV 75
R + LA +V +L SL +L ++G++G V + + F++
Sbjct 9 RSKYFGLATIFIVSILR--YSLCELTVFSIGDWGEKTECLVNVTTKMGSLESAMNPKFII 66
Query 76 SPGSNF-PNGVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDL 134
S G NF GV D W E+ E+ + K S + + LG DW G+ TA +RT+
Sbjct 67 SVGDNFYQRGVKSVEDAAWTEILEEPFG--KLSKHLKVHSCLGDHDWRGSTTAQIDRTNY 124
Query 135 VYGDGSSEEAVSNSPKWTLPNWWYHYAVHFTA--STGSAFLSSGHKDMSVGLVF 186
+N+ +W LP +W++ V FT+ S S F SG ++S F
Sbjct 125 -----------ANNTRWYLPGYWWYEKVTFTSEVSLPSLFEKSGLGEISANNSF 167
> xla:444356 MGC82831 protein; K14379 tartrate-resistant acid
phosphatase type 5 [EC:3.1.3.2]
Length=325
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/136 (30%), Positives = 67/136 (49%), Gaps = 21/136 (15%)
Query 33 IPASLAQLRFAALGNYG------FDISGQKEVAETLKKTAAEERISFLVSPGSNF-PNGV 85
+P LRF ALG++G F Q+ VAE + KT A+ F++S G NF +GV
Sbjct 20 VPHEEPSLRFVALGDWGGLPLPPFTTRQQELVAEEMSKTVAKLGADFILSLGDNFYYDGV 79
Query 86 SGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAV 145
+ +D ++ FE VYS E +++P+ + G D GN +A T++
Sbjct 80 TDESDPRFKFTFESVYSAES-LVKLPWYILAGNHDHKGNVSAQIAYTNV----------- 127
Query 146 SNSPKWTLPNWWYHYA 161
S +W P+++Y A
Sbjct 128 --STRWNYPDYYYDLA 141
> xla:432322 acp5, MGC78938; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=326
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 47/168 (27%), Positives = 77/168 (45%), Gaps = 32/168 (19%)
Query 33 IPASLAQLRFAALGNYG------FDISGQKEVAETLKKTAAEERISFLVSPGSNF-PNGV 85
+P LRF ALG++G + Q+ VAE + KT A+ F++S G NF +GV
Sbjct 21 VPRKDPTLRFVALGDWGGLPLPPYTTRQQELVAEEMGKTVAKLGADFILSLGDNFYYDGV 80
Query 86 SGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAV 145
+ +D ++ FE VYS E ++ P+ + G D GN +A T++
Sbjct 81 TDVSDPRFKITFESVYSSES-LIKHPWYILAGNHDHKGNVSAQIAYTNV----------- 128
Query 146 SNSPKWTLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLS 193
S +W P+++Y A S +++V L+ +DT L
Sbjct 129 --STRWNYPDYYYDLAFTIPGS-----------NVTVRLLMLDTVQLC 163
> dre:406801 acp5a, acp5, sb:cb576, wu:fb19f01, wu:fb30b03, wu:fi14e01,
wu:fj66f03, zgc:63825; acid phosphatase 5a, tartrate
resistant (EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=339
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 74/163 (45%), Gaps = 33/163 (20%)
Query 38 AQLRFAALGNYG------FDISGQKEVAETLKKTAAEERISFLVSPGSNFP-NGVSGAND 90
+ +RF LG++G + + A + KTA++ F+++ G NF GV+ ND
Sbjct 34 SSIRFLVLGDWGGLPNPPYVTPIETATARMMAKTASQMGADFILAVGDNFYYKGVTDVND 93
Query 91 TKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAVSNSPK 150
++ E FEDVY+ + SL +P+ + G D GN A E + S +
Sbjct 94 PRFQETFEDVYTQD--SLNIPWYVIAGNHDHVGNVKAQIEYS-------------QRSKR 138
Query 151 WTLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLS 193
W P +Y+Y ++F D ++ ++ +DT +L
Sbjct 139 WNFP--YYYYEMNFRIP---------RTDSTLTIIMLDTVLLC 170
> cel:F02E9.7 hypothetical protein; K14379 tartrate-resistant
acid phosphatase type 5 [EC:3.1.3.2]
Length=419
Score = 52.8 bits (125), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 58/115 (50%), Gaps = 16/115 (13%)
Query 55 QKEVAETLKKTAAEERISFLVSPGSN-FPNGVSGANDTKWDEMFEDVYSDEKGSLRMPFL 113
Q EV +T+ A E + +++ G N + G + D +++ FE+VY++ SL++P+L
Sbjct 100 QNEVKQTMASLADEHSVQMILNMGDNIYFTGPTDEFDPRFESRFENVYTNP--SLQVPWL 157
Query 114 TVLGAEDWSGNYTAMRERTDLVYGDGSSEEAVSNSPKWTLPNWWYHYAVHFTAST 168
T+ G D GN TA E T +S KW P+ +Y +V F ++
Sbjct 158 TIAGNHDHFGNVTAEIEYT-------------KHSKKWYFPSLYYKKSVEFNGTS 199
> hsa:54 ACP5, MGC117378, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=325
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 47/166 (28%), Positives = 76/166 (45%), Gaps = 42/166 (25%)
Query 40 LRFAALGNYG----------FDISGQKEVAETLKKTAAEERISFLVSPGSNFP-NGVSGA 88
LRF A+G++G +++ KE+A T++ A+ F++S G NF GV
Sbjct 26 LRFVAVGDWGGVPNAPFHTAREMANAKEIARTVQILGAD----FILSLGDNFYFTGVQDI 81
Query 89 NDTKWDEMFEDVYSDEKGSLR-MPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAVSN 147
ND ++ E FEDV+SD SLR +P+ + G D GN +A + +
Sbjct 82 NDKRFQETFEDVFSDR--SLRKVPWYVLAGNHDHLGNVSAQIAYSKI------------- 126
Query 148 SPKWTLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLS 193
S +W P+ +Y +HF ++SV + +DT L
Sbjct 127 SKRWNFPSPFYR--LHFKIP---------QTNVSVAIFMLDTVTLC 161
> mmu:11433 Acp5, TRACP, TRAP; acid phosphatase 5, tartrate resistant
(EC:3.1.3.2); K14379 tartrate-resistant acid phosphatase
type 5 [EC:3.1.3.2]
Length=327
Score = 50.1 bits (118), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 48/183 (26%), Positives = 84/183 (45%), Gaps = 43/183 (23%)
Query 40 LRFAALGNYG----------FDISGQKEVAETLKKTAAEERISFLVSPGSNFP-NGVSGA 88
LRF A+G++G +++ KE+A T++ A+ F++S G NF GV A
Sbjct 28 LRFVAVGDWGGVPNAPFHTAREMANAKEIARTVQTMGAD----FIMSLGDNFYFTGVHDA 83
Query 89 NDTKWDEMFEDVYSDEKGSLR-MPFLTVLGAEDWSGNYTAMRERTDLVYGDGSSEEAVSN 147
+D ++ E FEDV+SD +LR +P+ + G D GN +A + +
Sbjct 84 SDKRFQETFEDVFSDR--ALRNIPWYVLAGNHDHLGNVSAQIAYSKI------------- 128
Query 148 SPKWTLPNWWYHYAVHFTASTGSAFLSSGHKDMSVGLVFVDTWVLS-SAFPFGSVTEKAW 206
S +W P+ +Y + +++V + +DT +L ++ F S K
Sbjct 129 SKRWNFPSPYYRLRFKIPRT-----------NITVAIFMLDTVMLCGNSDDFASQQPKMP 177
Query 207 KDL 209
+DL
Sbjct 178 RDL 180
> ath:AT2G01890 PAP8; PAP8 (PURPLE ACID PHOSPHATASE 8); acid phosphatase/
protein serine/threonine phosphatase
Length=335
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 33/107 (30%), Positives = 55/107 (51%), Gaps = 4/107 (3%)
Query 23 AAAILVGLLEIPASLAQLRFAALGNYGFDIS-GQKEVAETLKKTAAEERISFLVSPGSNF 81
+ A L ++ P L F +G++G S Q +VA + K + I FL+S G NF
Sbjct 28 STAELPRFVQPPEPDGSLSFLVVGDWGRRGSYNQSQVALQMGKIGKDLNIDFLISTGDNF 87
Query 82 -PNGVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTA 127
+G+ D+++ + F ++Y+ SL+ P+ VLG D+ GN A
Sbjct 88 YDDGIISPYDSQFQDSFTNIYT--ATSLQKPWYNVLGNHDYRGNVYA 132
> dre:436725 acp5b, zgc:92339; acid phosphatase 5b, tartrate resistant;
K14379 tartrate-resistant acid phosphatase type 5
[EC:3.1.3.2]
Length=327
Score = 45.4 bits (106), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 37/149 (24%), Positives = 68/149 (45%), Gaps = 28/149 (18%)
Query 24 AAILVGLLEIPASLAQ-------LRFAALGNYG------FDISGQKEVAETLKKTAAEER 70
A +L+ L+ ++ +Q LRF +G++G F + + A+ L + A
Sbjct 6 AFLLISCLQTFSTASQTNQQASSLRFVGIGDWGGRPSYPFYTPHEADTAKELARVAQSSG 65
Query 71 ISFLVSPGSNF-PNGVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMR 129
+ F++S G NF +GV +DT++ +E V+S + +P+ + G D GN +A
Sbjct 66 LDFVLSLGDNFYYDGVKDVDDTRFKFSYEQVFS-HPALMTIPWYLIAGNHDHRGNVSAQ- 123
Query 130 ERTDLVYGDGSSEEAVSNSPKWTLPNWWY 158
+ Y S S +W P+ +Y
Sbjct 124 ----IAYS--------SRSERWIYPDLYY 140
> ath:AT1G25230 purple acid phosphatase family protein
Length=339
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/105 (28%), Positives = 52/105 (49%), Gaps = 4/105 (3%)
Query 25 AILVGLLEIPASLAQLRFAALGNYG-FDISGQKEVAETLKKTAAEERISFLVSPGSN-FP 82
A L + P + F +G++G + Q +VA + + E I+F+VS G N +
Sbjct 31 AELATVQHAPNPDGSISFLVIGDWGRHGLYNQSQVALQMGRIGEEMDINFVVSTGDNIYD 90
Query 83 NGVSGANDTKWDEMFEDVYSDEKGSLRMPFLTVLGAEDWSGNYTA 127
NG+ +D + F ++Y+ SL+ P+ VLG D+ G+ A
Sbjct 91 NGMKSIDDPAFQLSFSNIYTSP--SLQKPWYLVLGNHDYRGDVEA 133
> ath:AT2G01880 PAP7; PAP7 (PURPLE ACID PHOSPHATASE 7); acid phosphatase/
protein serine/threonine phosphatase
Length=328
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 28/95 (29%), Positives = 46/95 (48%), Gaps = 4/95 (4%)
Query 40 LRFAALGNYGFDIS-GQKEVAETLKKTAAEERISFLVSPGSNF-PNGVSGANDTKWDEMF 97
L F +G++G Q VA + + I F++S G NF +G+ G ND ++ F
Sbjct 41 LSFLVIGDWGRKGGFNQSLVAHQMGVVGEKLDIDFVISVGDNFYDDGLKGVNDPSFEASF 100
Query 98 EDVYSDEKGSLRMPFLTVLGAEDWSGNYTAMRERT 132
+Y+ SL+ + +VLG D+ GN A +
Sbjct 101 SHIYTHP--SLQKQWYSVLGNHDYRGNVEAQLSKV 133
> tgo:TGME49_097650 serine/threonine protein phosphatase, putative
(EC:3.1.3.2)
Length=679
Score = 35.0 bits (79), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 37/160 (23%), Positives = 56/160 (35%), Gaps = 54/160 (33%)
Query 42 FAALGNYGFDISGQKEVAETLKKTAAEERISFLVSPGSN-FPNGVSGANDTKWDEMFEDV 100
FA++G+ G S Q + +L + I F+ G N +P+GV+ A D W FE
Sbjct 151 FASIGDTGAANSNQAKCGISLAALSVALDIKFVNLLGDNLYPHGVTSAVDPLWQSAFE-- 208
Query 101 YSDEKGSLRMPFLTVLGAEDWSGNYTAMRER-----------TDLVYGDGSSEEAVS--- 146
+ F VLG D+ + A +R T+ DGS +
Sbjct 209 ---------VAFFPVLGNHDYHLDPYAQIDRCIRLGNEHLNFTEKEKTDGSRPQLSKTYQ 259
Query 147 ----------------------------NSPKWTLPNWWY 158
+ P+W LPN+WY
Sbjct 260 MQAMLPWMKIHHEKFRYSFDQTTKMHGLDGPRWRLPNFWY 299
> tgo:TGME49_028170 serine/threonine protein phosphatase, putative
(EC:3.1.3.2 3.2.1.3)
Length=1491
Score = 34.7 bits (78), Expect = 0.24, Method: Composition-based stats.
Identities = 33/146 (22%), Positives = 58/146 (39%), Gaps = 15/146 (10%)
Query 57 EVAETLKKTAAEERISFLVSPGS--NFPNGVSGANDTKWDEMFEDVYSDEKGSLRMPFLT 114
+ L K AEE+ ++ G P +S A D ++ + + D++ + L +P+L
Sbjct 424 QTVSALAKWHAEEKADAVLGLGDFLGIPGPLS-ARDERFTKRWYDIFVKD-AKLDIPWLM 481
Query 115 VLGAEDWSGNYTAMRERTDLVYGDGSSEEAVSNSPKWTLPNWWYHYAVHFTASTGSAFLS 174
LG E+ N +A P W +PN Y F+ S A +
Sbjct 482 TLGEEEALVNPSA-----------SVRHHYTGEHPNWYMPNDAYTATFSFSTSMTMANGT 530
Query 175 SGHKDMSVGLVFVDTWVLSSAFPFGS 200
H+ + ++ V+TW L P +
Sbjct 531 IQHEAFNATVINVNTWNLFVGNPIAN 556
> cel:T12E12.6 hypothetical protein
Length=641
Score = 31.2 bits (69), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query 16 RVRPVALAA-AILVGLLEIPASLAQLRFAALGNY--GFDISGQKEVAETLKKTAAEERIS 72
RV ALA+ IL GL+++P L ++ F + Y G + G V+ETL + + +
Sbjct 237 RVLDCALASFEILAGLIDVPLPLNKIDFILVPEYDGGMENWGHITVSETLATSGDDAHLI 296
Query 73 FLVS 76
+L++
Sbjct 297 YLIA 300
> tgo:TGME49_009190 ABC transporter, putative (EC:3.6.3.44)
Length=798
Score = 30.4 bits (67), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 1/65 (1%)
Query 47 NYGFDISGQKEVAETLKKTAAEERISFLVSPGS-NFPNGVSGANDTKWDEMFEDVYSDEK 105
N G +I KE ET TAA + +S + PGS G SGA + ++ ++
Sbjct 506 NEGREIGESKETPETTGDTAALKNVSLVFPPGSVTAVTGASGAGKSTLLKLLTQELRPQQ 565
Query 106 GSLRM 110
G++ +
Sbjct 566 GTVTL 570
> tgo:TGME49_078110 1,3-beta-glucan synthase component domain-containing
protein (EC:2.4.1.34); K00706 1,3-beta-glucan synthase
[EC:2.4.1.34]
Length=2321
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 19/38 (50%), Gaps = 3/38 (7%)
Query 156 WWYHYAVHFTASTGSA---FLSSGHKDMSVGLVFVDTW 190
W ++Y HF+ F+ SG+ D GL F+D W
Sbjct 2183 WQFYYFTHFSIGLEMMMLLFIYSGYCDFDAGLYFLDVW 2220
> cel:F54E7.3 par-3; abnormal embryonic PARtitioning of cytoplasm
family member (par-3); K04237 partitioning defective protein
3
Length=1379
Score = 30.0 bits (66), Expect = 6.5, Method: Composition-based stats.
Identities = 20/59 (33%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 55 QKEVAETLKKTAAEERISFLVSPGSNFP-NGVSGANDTKWDEMFEDVYSDEKGSLRMPF 112
Q E+ E LK+T E+I FLVS S S +++ K +E V +EK ++P
Sbjct 575 QSEIVEKLKETMVGEKIKFLVSRVSQSAIMSTSASSENKENEETLKVVEEEKIPQKLPL 633
Lambda K H
0.316 0.131 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6494887820
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40