bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_2075_orf3
Length=226
Score E
Sequences producing significant alignments: (Bits) Value
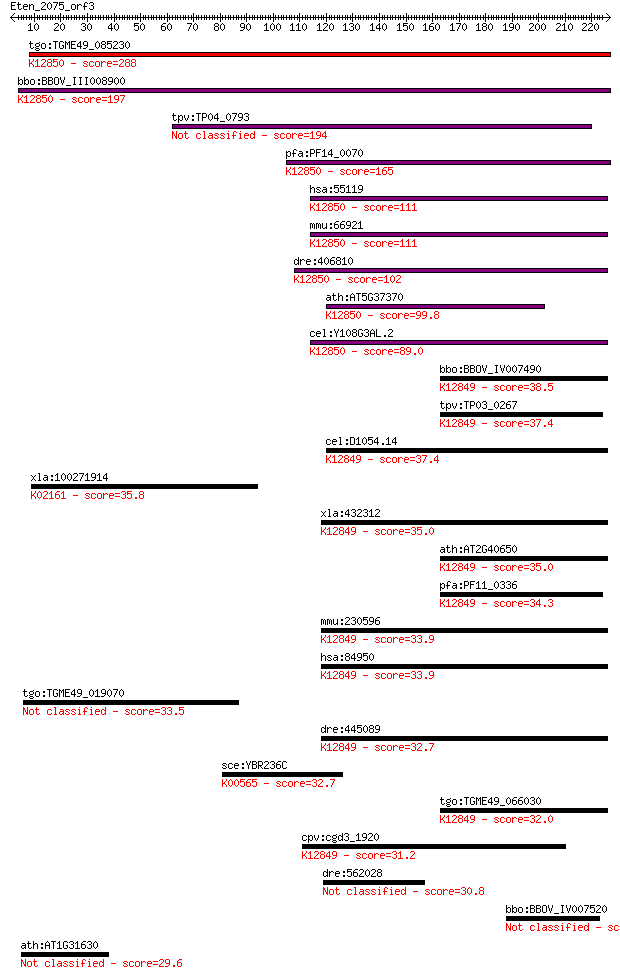
tgo:TGME49_085230 PRP38 family domain-containing protein ; K12... 288 1e-77
bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre... 197 2e-50
tpv:TP04_0793 hypothetical protein 194 3e-49
pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-m... 165 1e-40
hsa:55119 PRPF38B, FLJ10330, MGC163218, MGC41809, NET1, RP11-2... 111 2e-24
mmu:66921 Prpf38b, 1110021E09Rik, AU018955; PRP38 pre-mRNA pro... 111 2e-24
dre:406810 prpf38b, wu:fc43b05, zgc:63496; PRP38 pre-mRNA proc... 102 1e-21
ath:AT5G37370 ATSRL1; ATSRL1; binding; K12850 pre-mRNA-splicin... 99.8 8e-21
cel:Y108G3AL.2 hypothetical protein; K12850 pre-mRNA-splicing ... 89.0 1e-17
bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849 ... 38.5 0.018
tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing f... 37.4 0.038
cel:D1054.14 hypothetical protein; K12849 pre-mRNA-splicing fa... 37.4 0.040
xla:100271914 bcl2, bcl-2, xBcl-2, xbcl2; B-cell CLL/lymphoma ... 35.8 0.11
xla:432312 prpf38a, MGC116433; PRP38 pre-mRNA processing facto... 35.0 0.23
ath:AT2G40650 pre-mRNA splicing factor PRP38 family protein; K... 35.0 0.23
pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-m... 34.3 0.32
mmu:230596 Prpf38a, 2410002M20Rik; PRP38 pre-mRNA processing f... 33.9 0.42
hsa:84950 PRPF38A, FLJ14936, MGC3320, Prp38, RP5-965L7.1; PRP3... 33.9 0.54
tgo:TGME49_019070 cAMP-dependent protein kinase regulatory sub... 33.5 0.67
dre:445089 prpf38a, MGC173831, fa04h11, fc83e08, pmp22b, wu:fa... 32.7 0.99
sce:YBR236C ABD1; Abd1p (EC:2.1.1.56); K00565 mRNA (guanine-N7... 32.7 1.1
tgo:TGME49_066030 PRP38 family domain-containing protein ; K12... 32.0 1.8
cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family m... 31.2 3.5
dre:562028 zcchc24, fc45d05, wu:fc45d05, zgc:158323; zinc fing... 30.8 4.5
bbo:BBOV_IV007520 23.m06029; S1/P1 nuclease 30.4 4.9
ath:AT1G31630 AGL86; AGL86 (AGAMOUS-LIKE 86); DNA binding / tr... 29.6 8.9
> tgo:TGME49_085230 PRP38 family domain-containing protein ; K12850
pre-mRNA-splicing factor 38B
Length=614
Score = 288 bits (737), Expect = 1e-77, Method: Compositional matrix adjust.
Identities = 141/224 (62%), Positives = 169/224 (75%), Gaps = 10/224 (4%)
Query 8 APLFYGATSAATAAVAAAAVAPIAAAAAAPVAVA-PLAAAAADVGAAD--KDKGDEEEAI 64
AP FYGA +A AA+ + +A P VA P A VGA D +D GD A+
Sbjct 39 APRFYGAPAAPPAALPHR----LGVSAGTPAVVAAPGPPATTAVGAVDAEEDSGDTW-AV 93
Query 65 ARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAAEEETQ--RKNMVQMTDSTTYNVNQ 122
RCHLHTKPQL CKFCRKYKSF QQ+G+LQ+R + +E+ ++NMV+MTDSTTYNVN
Sbjct 94 NRCHLHTKPQLHCKFCRKYKSFAQQLGILQKRELKQQEDDDGMKRNMVEMTDSTTYNVNA 153
Query 123 LLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLFTLK 182
LLR NI+SSEYFKSLH+ K+ EVVDE+ +A HAEPYCSGS+RAPSTLFCCLYKLFT+K
Sbjct 154 LLRSNILSSEYFKSLHELKTVPEVVDEITQYAQHAEPYCSGSSRAPSTLFCCLYKLFTMK 213
Query 183 LTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYFL 226
LT KQ+ LL++ +SPYVRCTGFLYLRYVHP ++LWKWYE YFL
Sbjct 214 LTTKQLEQLLDYSDSPYVRCTGFLYLRYVHPPEKLWKWYEQYFL 257
> bbo:BBOV_III008900 17.m07776; PRP38 family protein; K12850 pre-mRNA-splicing
factor 38B
Length=483
Score = 197 bits (502), Expect = 2e-50, Method: Compositional matrix adjust.
Identities = 104/223 (46%), Positives = 144/223 (64%), Gaps = 17/223 (7%)
Query 4 HAGVAPLFYGATSAATAAVAAAAVAPIAAAAAAPVAVAPLAAAAADVGAADKDKGDEEEA 63
+ G+ P+ AT T+ AAA P P +V+ D E+
Sbjct 82 NVGIQPI---ATELGTSDNIAAASVP-GHMELPPTSVSYTTHTE-----------DLLES 126
Query 64 IARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAAEEETQRKNMVQMTDSTTYNVNQL 123
+ CH+HTKP L+CKFCRKYKS ++ L Q + ++E T+R + + MT+S+TYN+N L
Sbjct 127 VQTCHMHTKPDLNCKFCRKYKSAVHELSRLSQ--LRSQEATERPDQLDMTNSSTYNMNTL 184
Query 124 LRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLFTLKL 183
L NI++S+Y+KSL S + ++DELA +ADH EPYC +TR PSTLFCCL+KLFTL+L
Sbjct 185 LLNNILNSDYYKSLSTMTSHHSIIDELAQYADHVEPYCKTATRVPSTLFCCLHKLFTLRL 244
Query 184 TEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYFL 226
TE+QM L++ +SPY RC GFLYLR+V P+DQLW W+EPY +
Sbjct 245 TERQMEDLIDCTKSPYPRCCGFLYLRFVLPSDQLWAWFEPYLM 287
> tpv:TP04_0793 hypothetical protein
Length=239
Score = 194 bits (492), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 90/158 (56%), Positives = 117/158 (74%), Gaps = 2/158 (1%)
Query 62 EAIARCHLHTKPQLSCKFCRKYKSFTQQMGVLQQRAIAAEEETQRKNMVQMTDSTTYNVN 121
E++ RCHLHTKP+L+CKFCRK+KS ++ + ++ + + N + MT+S TYN+N
Sbjct 81 ESVQRCHLHTKPELNCKFCRKFKSSVHEISK-IAQKKSSSSKEDKANQIPMTNSVTYNMN 139
Query 122 QLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLFTL 181
LLR NI+SSEY+KSL K+F +V DEL FA H+EPYCS +TRAPST+FCCLY+ L
Sbjct 140 DLLRSNILSSEYYKSL-SVKNFYQVFDELVQFATHSEPYCSTATRAPSTIFCCLYRFLVL 198
Query 182 KLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWK 219
KLTEKQM+ LL + +SPY RC GFLYLRYV P D+LW
Sbjct 199 KLTEKQMNFLLENVKSPYARCCGFLYLRYVLPPDKLWN 236
> pfa:PF14_0070 pre-mRNA splicing factor, putative; K12850 pre-mRNA-splicing
factor 38B
Length=690
Score = 165 bits (417), Expect = 1e-40, Method: Composition-based stats.
Identities = 77/122 (63%), Positives = 98/122 (80%), Gaps = 0/122 (0%)
Query 105 QRKNMVQMTDSTTYNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGS 164
++KN ++MT++TTYNVN LLR NI+SSEYFKSL K+F EVVDE+ ++ADH EPYC GS
Sbjct 168 EKKNCLEMTNTTTYNVNTLLRNNILSSEYFKSLIPIKTFKEVVDEIHSYADHVEPYCIGS 227
Query 165 TRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPY 224
RAPSTLFCCLYK FT++L+EKQ+ L+ +++S Y+R GFLYLRYVH LW W+EPY
Sbjct 228 NRAPSTLFCCLYKFFTMQLSEKQLKSLIENKDSCYIRACGFLYLRYVHSPANLWMWFEPY 287
Query 225 FL 226
L
Sbjct 288 LL 289
> hsa:55119 PRPF38B, FLJ10330, MGC163218, MGC41809, NET1, RP11-293A10.1;
PRP38 pre-mRNA processing factor 38 (yeast) domain
containing B; K12850 pre-mRNA-splicing factor 38B
Length=546
Score = 111 bits (278), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 75/131 (57%), Gaps = 19/131 (14%)
Query 114 DSTTYNVNQLLRGNIMSSEYFK-SLHQFKSFNEVVDELAAFADHAEPYCSGSTRA----- 167
+ T N+N ++ NI+SS YFK L++ K+++EVVDE+ H EP+ GS +
Sbjct 52 NEKTMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTG 111
Query 168 -------------PSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPA 214
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY P
Sbjct 112 MCGGVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 171
Query 215 DQLWKWYEPYF 225
LW W+E +
Sbjct 172 TDLWDWFESFL 182
> mmu:66921 Prpf38b, 1110021E09Rik, AU018955; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing B; K12850 pre-mRNA-splicing
factor 38B
Length=542
Score = 111 bits (277), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 54/131 (41%), Positives = 75/131 (57%), Gaps = 19/131 (14%)
Query 114 DSTTYNVNQLLRGNIMSSEYFK-SLHQFKSFNEVVDELAAFADHAEPYCSGSTRA----- 167
+ T N+N ++ NI+SS YFK L++ K+++EVVDE+ H EP+ GS +
Sbjct 53 NEKTMNLNPMILTNILSSPYFKVQLYELKTYHEVVDEIYFKVTHVEPWEKGSRKTAGQTG 112
Query 168 -------------PSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPA 214
ST FC LYKLFTLKLT KQ+ L+ H +SPY+R GF+Y+RY P
Sbjct 113 MCGGVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHTDSPYIRALGFMYIRYTQPP 172
Query 215 DQLWKWYEPYF 225
LW W+E +
Sbjct 173 TDLWDWFESFL 183
> dre:406810 prpf38b, wu:fc43b05, zgc:63496; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing B; K12850 pre-mRNA-splicing
factor 38B
Length=501
Score = 102 bits (253), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 52/137 (37%), Positives = 76/137 (55%), Gaps = 19/137 (13%)
Query 108 NMVQMTDSTTYNVNQLLRGNIMSSEYFK-SLHQFKSFNEVVDELAAFADHAEPYCSGSTR 166
N+ + T N+N ++ N++SS YFK L++ K+++EVVDE+ +H EP+ GS +
Sbjct 24 NLPLWGNEKTMNLNPMILTNVLSSPYFKVQLYELKTYHEVVDEIYFKVNHVEPWEKGSRK 83
Query 167 A------------------PSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYL 208
ST FC LYKLFTLKLT KQ+ L+ H +SP +R GF+Y+
Sbjct 84 TAGQTGMCGGVRGVGTGGIVSTAFCLLYKLFTLKLTRKQVMGLITHSDSPDIRALGFMYI 143
Query 209 RYVHPADQLWKWYEPYF 225
RY P L WY+ +
Sbjct 144 RYTQPPPDLVDWYDEFL 160
> ath:AT5G37370 ATSRL1; ATSRL1; binding; K12850 pre-mRNA-splicing
factor 38B
Length=111
Score = 99.8 bits (247), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 60/82 (73%), Gaps = 0/82 (0%)
Query 120 VNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLF 179
+ ++L NI+SS+YFK L+ K+++EV+DE+ +H EP+ G+ R PST +C LYK F
Sbjct 15 LEKVLSMNILSSDYFKELYGLKTYHEVIDEIYNQVNHVEPWMGGNCRGPSTAYCLLYKFF 74
Query 180 TLKLTEKQMHMLLNHRESPYVR 201
T+KLT KQMH LL H +SPY+R
Sbjct 75 TMKLTVKQMHGLLKHTDSPYIR 96
> cel:Y108G3AL.2 hypothetical protein; K12850 pre-mRNA-splicing
factor 38B
Length=327
Score = 89.0 bits (219), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 69/131 (52%), Gaps = 19/131 (14%)
Query 114 DSTTYNVNQLLRGNIMSSEYFKS-LHQFKSFNEVVDELAAFADHAEPYCSGSTR------ 166
+ T N+N L+ NI S Y+K+ L + +F +++++ H EP+ G+ R
Sbjct 60 NKVTMNLNTLVLENIRESYYYKNNLVEIDNFQTLIEQIFYQVKHLEPWEKGTRRLQGMTG 119
Query 167 ------------APSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPA 214
S+ +C LY+LF LK++ KQ+ +LN R+S Y+R GF+Y+RY P
Sbjct 120 MCGGVRGVGAGGVVSSAYCLLYRLFNLKISRKQLISMLNSRQSVYIRGLGFMYIRYTQPP 179
Query 215 DQLWKWYEPYF 225
LW W EPY
Sbjct 180 ADLWYWLEPYL 190
> bbo:BBOV_IV007490 23.m06033; pre-mRNA splicing factor; K12849
pre-mRNA-splicing factor 38A
Length=202
Score = 38.5 bits (88), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 18/63 (28%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G R PS C + KL ++ + + + + E Y+R G Y+R V A Q+++ E
Sbjct 62 GGNRQPSPFLCLVLKLLQIQPEIEIIQEYIRNEEFKYLRALGIYYMRLVGNAVQIYQNLE 121
Query 223 PYF 225
P +
Sbjct 122 PVY 124
> tpv:TP03_0267 hypothetical protein; K12849 pre-mRNA-splicing
factor 38A
Length=193
Score = 37.4 bits (85), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 31/61 (50%), Gaps = 0/61 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G R PS C + K+ ++ + +H + + + Y+R G Y+R V A ++++ E
Sbjct 62 GGNRQPSPFICLVLKMLQIQPEMEIVHEYIKNEDYKYLRALGIYYMRLVGTAVEVYRTLE 121
Query 223 P 223
P
Sbjct 122 P 122
> cel:D1054.14 hypothetical protein; K12849 pre-mRNA-splicing
factor 38A
Length=320
Score = 37.4 bits (85), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 24/106 (22%), Positives = 49/106 (46%), Gaps = 2/106 (1%)
Query 120 VNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYKLF 179
V +++R I S Y+K H F E+V + + +G+ + P+ C K+
Sbjct 21 VEKIIRQRIYDSMYWKE-HCFALTAELVVDKGMDLRYIGGIYAGNIK-PTPFLCLALKML 78
Query 180 TLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
++ + + + E Y+R G +YLR + +++K+ EP +
Sbjct 79 QIQPDKDIVLEFIQQEEFKYIRALGAMYLRLTFDSTEIYKYLEPLY 124
> xla:100271914 bcl2, bcl-2, xBcl-2, xbcl2; B-cell CLL/lymphoma
2; K02161 apoptosis regulator BCL-2
Length=294
Score = 35.8 bits (81), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 28/90 (31%), Positives = 43/90 (47%), Gaps = 5/90 (5%)
Query 9 PLFYGATSAATAAVAAAAVAPIAAAAAAP--VAVAPLAAAAADVGAADKDKGDEE--EAI 64
P+ + + A +A+AV+P+A P + V P A+ AAD D D GDE + +
Sbjct 87 PITFVGNAPAVPRRSASAVSPLAELNVEPRDLNVNPDASRAADGDNNDADGGDEAVMQPV 146
Query 65 ARCHLHTKPQLSCKFCRKY-KSFTQQMGVL 93
L T + +F R Y + F Q G+L
Sbjct 147 PSAVLQTLSRAGDEFSRLYQQDFRQISGLL 176
> xla:432312 prpf38a, MGC116433; PRP38 pre-mRNA processing factor
38 domain containing A; K12849 pre-mRNA-splicing factor
38A
Length=312
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Query 118 YNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYK 177
Y V +++R I S+Y+K + VVD+ Y G P+ C K
Sbjct 19 YLVEKIIRTRIYESKYWKEECFGLTAELVVDKAMELKFVGGVY--GGNIKPTPFLCLTLK 76
Query 178 LFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
+ ++ + + + + + YVR G LY+R A +K+ EP +
Sbjct 77 MLQIQPEKDIIVEFIKNEDFKYVRALGALYMRLTGTATDCYKYLEPLY 124
> ath:AT2G40650 pre-mRNA splicing factor PRP38 family protein;
K12849 pre-mRNA-splicing factor 38A
Length=355
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 14/63 (22%), Positives = 31/63 (49%), Gaps = 0/63 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G +R P+ C + K+ ++ ++ + + + + YVR G YLR ++++ E
Sbjct 62 GGSRKPTPFLCLILKMLQIQPEKEIVVEFIKNDDYKYVRILGAFYLRLTGTDVDVYRYLE 121
Query 223 PYF 225
P +
Sbjct 122 PLY 124
> pfa:PF11_0336 pre-mRNA splicing factor, putative; K12849 pre-mRNA-splicing
factor 38A
Length=386
Score = 34.3 bits (77), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 14/61 (22%), Positives = 30/61 (49%), Gaps = 0/61 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G R P+ C + KL ++ + ++ + + + Y+R G YLR + + +++ E
Sbjct 62 GGNRKPTRFLCLILKLLQIQPDKDIIYEYIKNEDFVYLRALGIFYLRLIGKSLEVYNHLE 121
Query 223 P 223
P
Sbjct 122 P 122
> mmu:230596 Prpf38a, 2410002M20Rik; PRP38 pre-mRNA processing
factor 38 (yeast) domain containing A; K12849 pre-mRNA-splicing
factor 38A
Length=312
Score = 33.9 bits (76), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Query 118 YNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYK 177
Y V +++R I S+Y+K + VVD+ Y G P+ C K
Sbjct 19 YLVEKIIRTRIYESKYWKEECFGLTAELVVDKAMELKFVGGVY--GGNIKPTPFLCLTLK 76
Query 178 LFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
+ ++ + + + + + YVR G LY+R A +K+ EP +
Sbjct 77 MLQIQPEKDIIVEFIKNEDFKYVRMLGALYMRLTGTAIDCYKYLEPLY 124
> hsa:84950 PRPF38A, FLJ14936, MGC3320, Prp38, RP5-965L7.1; PRP38
pre-mRNA processing factor 38 (yeast) domain containing
A; K12849 pre-mRNA-splicing factor 38A
Length=312
Score = 33.9 bits (76), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Query 118 YNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYK 177
Y V +++R I S+Y+K + VVD+ Y G P+ C K
Sbjct 19 YLVEKIIRTRIYESKYWKEECFGLTAELVVDKAMELRFVGGVY--GGNIKPTPFLCLTLK 76
Query 178 LFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
+ ++ + + + + + YVR G LY+R A +K+ EP +
Sbjct 77 MLQIQPEKDIIVEFIKNEDFKYVRMLGALYMRLTGTAIDCYKYLEPLY 124
> tgo:TGME49_019070 cAMP-dependent protein kinase regulatory subunit,
putative (EC:3.6.3.14 2.7.10.2 2.7.11.12)
Length=2634
Score = 33.5 bits (75), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 36/82 (43%), Gaps = 2/82 (2%)
Query 6 GVAPLFYGATSAATAAVAAAAVAPIAAAAAAPVAVAPLAAAAADVGAADKDKGDEE-EAI 64
G P G + A++A + +AP+ A A +A P AA+ GA + D D E E++
Sbjct 904 GQPPRSLGRLAKRNASLATS-IAPVKMAEVADLAENPAVAASTSSGAGEGDLSDSETESV 962
Query 65 ARCHLHTKPQLSCKFCRKYKSF 86
+ L K KY S
Sbjct 963 TNASYSPRTALKHKILDKYNSL 984
> dre:445089 prpf38a, MGC173831, fa04h11, fc83e08, pmp22b, wu:fa04h11,
wu:fc83e08, zgc:92059; PRP38 pre-mRNA processing factor
38 (yeast) domain containing A; K12849 pre-mRNA-splicing
factor 38A
Length=313
Score = 32.7 bits (73), Expect = 0.99, Method: Compositional matrix adjust.
Identities = 24/108 (22%), Positives = 46/108 (42%), Gaps = 2/108 (1%)
Query 118 YNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPSTLFCCLYK 177
Y V +++R I S+Y+K + VVD+ Y G P+ C K
Sbjct 19 YLVEKIIRTRIYESKYWKEECFGLTAELVVDKAMELKFVGGVY--GGNVKPTPFLCLTLK 76
Query 178 LFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYEPYF 225
+ ++ + + + + + YVR G +Y+R + +K+ EP +
Sbjct 77 MLQIQPEKDIIVEFIKNEDFKYVRLLGAMYMRLTGTSVDCYKYLEPLY 124
> sce:YBR236C ABD1; Abd1p (EC:2.1.1.56); K00565 mRNA (guanine-N7-)-methyltransferase
[EC:2.1.1.56]
Length=436
Score = 32.7 bits (73), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 81 RKYKSFTQQMGVLQQRAIAAEEETQRKNMVQMTDSTTYNVNQLLR 125
R+++ + Q+ + +QRA EE +++ ++MT + + NV+Q++R
Sbjct 79 RRHERYDQEERLRKQRAQKLREEQLKRHEIEMTANRSINVDQIVR 123
> tgo:TGME49_066030 PRP38 family domain-containing protein ; K12849
pre-mRNA-splicing factor 38A
Length=524
Score = 32.0 bits (71), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/63 (25%), Positives = 28/63 (44%), Gaps = 0/63 (0%)
Query 163 GSTRAPSTLFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
G R P+ C + KL ++ + + + + Y+R G YLR V A +++ E
Sbjct 62 GGKRQPAPFLCLVLKLLQIQPEPEIILEFIKQEQFKYLRAVGAFYLRLVGRACEVYTHLE 121
Query 223 PYF 225
P
Sbjct 122 PLL 124
> cpv:cgd3_1920 possible PRP38 pre-mRNA splicing factor family
member ; K12849 pre-mRNA-splicing factor 38A
Length=261
Score = 31.2 bits (69), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 2/99 (2%)
Query 111 QMTDSTTYNVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADHAEPYCSGSTRAPST 170
Q + ++V+ +LR + SS Y+K F +E + + A D+ G R +
Sbjct 5 QNSHHKLFSVSSILRDRVFSSIYWKG-ECFALDSETILDKAVLLDYIGT-TYGGDRKATP 62
Query 171 LFCCLYKLFTLKLTEKQMHMLLNHRESPYVRCTGFLYLR 209
C L KL ++ + + + +N+ Y+ G +YLR
Sbjct 63 FLCLLVKLLQIRPSTEIVLEYINNPRFKYLTALGIVYLR 101
> dre:562028 zcchc24, fc45d05, wu:fc45d05, zgc:158323; zinc finger,
CCHC domain containing 24
Length=238
Score = 30.8 bits (68), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 119 NVNQLLRGNIMSSEYFKSLHQFKSFNEVVDELAAFADH 156
N QL R +++ +KS+ + S N +VD L + DH
Sbjct 71 NFFQLQRNEALNNGLYKSVSPYGSLNNIVDGLNSLTDH 108
> bbo:BBOV_IV007520 23.m06029; S1/P1 nuclease
Length=393
Score = 30.4 bits (67), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 22/35 (62%), Gaps = 3/35 (8%)
Query 188 MHMLLNHRESPYVRCTGFLYLRYVHPADQLWKWYE 222
++ +L+HRE Y TG L R HP D +W+W++
Sbjct 301 LNYILSHREIAYCEKTGLLIER--HP-DDVWRWHD 332
> ath:AT1G31630 AGL86; AGL86 (AGAMOUS-LIKE 86); DNA binding /
transcription factor
Length=339
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 17/33 (51%), Gaps = 0/33 (0%)
Query 5 AGVAPLFYGATSAATAAVAAAAVAPIAAAAAAP 37
AG APL A+ AVA AP+A A A P
Sbjct 189 AGAAPLAVAGAGASPLAVAGVGAAPLAVAGAGP 221
Lambda K H
0.321 0.129 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7504534908
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40