bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1937_orf1
Length=193
Score E
Sequences producing significant alignments: (Bits) Value
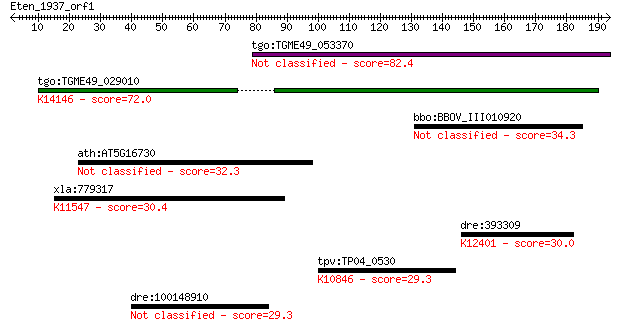
tgo:TGME49_053370 hypothetical protein 82.4 9e-16
tgo:TGME49_029010 hypothetical protein ; K14146 rhoptry neck p... 72.0 1e-12
bbo:BBOV_III010920 17.m07940; hypothetical protein 34.3 0.29
ath:AT5G16730 hypothetical protein 32.3 1.2
xla:779317 kntc2, MGC154845; HEC protein; K11547 kinetochore p... 30.4 3.6
dre:393309 ap4b1l, MGC63878, zgc:63878; adaptor-related protei... 30.0 4.8
tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision rep... 29.3 8.9
dre:100148910 hypothetical LOC100148910 29.3 9.1
> tgo:TGME49_053370 hypothetical protein
Length=1933
Score = 82.4 bits (202), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 65/117 (55%), Gaps = 2/117 (1%)
Query 79 SVSTSTTWSQVKAIIDKAHGPHSKVPLFLPSSSKCHCVRSNALGRCYIDILKTTEDLIFR 138
S S W + ++ GP +++P ++ +S C C+ C +D +K+T D+IFR
Sbjct 1304 STSRRLAWPYINKVLTAKSGPMTRMPSWVRTSDTCPCLDPEISEGCRMDSVKSTTDVIFR 1363
Query 139 LSLLLPAVLAEHI--KPKTKDLSTQAAALCAAAGVFVTAWQQQQVEAGLEHPNGRLA 193
LSLL+P V+ + + + T+ L AA CA+ G+FV++WQQ V G E G +A
Sbjct 1364 LSLLIPPVIEKFMSSQSSTRQLEPPFAAFCASVGIFVSSWQQHLVAGGHEEKGGNMA 1420
> tgo:TGME49_029010 hypothetical protein ; K14146 rhoptry neck
protein 4
Length=984
Score = 72.0 bits (175), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/107 (40%), Positives = 59/107 (55%), Gaps = 9/107 (8%)
Query 86 WSQVKAIIDKAHGPHSKVPLFLPSSSKCHCVRSNALGRCY--IDILKTTEDLIFRLSLLL 143
W +++ I KAHGP ++VP + P + C ALG Y ID+ K T D++FR++LL+
Sbjct 502 WPKMQNAISKAHGPLTRVPEWTPVTGSC------ALGDGYTDIDVTKATTDVLFRITLLI 555
Query 144 PAVLAEHIKPKTKDLSTQA-AALCAAAGVFVTAWQQQQVEAGLEHPN 189
+ + K QA ALC+AAG FV AWQ QQ LE P
Sbjct 556 LQQIRRKKTERGKLEDDQALVALCSAAGAFVDAWQHQQQALILEDPG 602
Score = 44.7 bits (104), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 30/64 (46%), Gaps = 0/64 (0%)
Query 10 RHVAEDVVPFLKRLSYSTPDAYGQVCQRLGWTTPRLPNTPEEAEAFVAALRDVRALQGPV 69
R + E FL+ LS P V + LGW +LP EE + F+ AL A+ G
Sbjct 340 RALKEQFFQFLQHLSADYPKQVQTVYEFLGWVADKLPENEEEVQMFIDALNTTEAMVGKA 399
Query 70 RRWL 73
RW+
Sbjct 400 ARWI 403
> bbo:BBOV_III010920 17.m07940; hypothetical protein
Length=1194
Score = 34.3 bits (77), Expect = 0.29, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 30/54 (55%), Gaps = 0/54 (0%)
Query 131 TTEDLIFRLSLLLPAVLAEHIKPKTKDLSTQAAALCAAAGVFVTAWQQQQVEAG 184
+T+DLI R+ +++ L + +T + + Q A LC+ A + V W+ Q+ G
Sbjct 644 STDDLIDRIHVMIGTWLDGYQGEETPEGNFQLATLCSTAAILVQQWRYMQLSQG 697
> ath:AT5G16730 hypothetical protein
Length=853
Score = 32.3 bits (72), Expect = 1.2, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 19/75 (25%)
Query 23 LSYSTPDAYGQVCQRLGWTTPRLPNTPEEAEAFVAALRDVRALQGPVRRWLADLGESVST 82
L S+P++ V +R +P+LP PE+++A VAA++ ES T
Sbjct 56 LDRSSPNSKSSVERR----SPKLPTPPEKSQARVAAVKGT---------------ESPQT 96
Query 83 STTWSQVKAIIDKAH 97
+T SQ+K + KA+
Sbjct 97 TTRLSQIKEDLKKAN 111
> xla:779317 kntc2, MGC154845; HEC protein; K11547 kinetochore
protein NDC80
Length=640
Score = 30.4 bits (67), Expect = 3.6, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 38/78 (48%), Gaps = 4/78 (5%)
Query 15 DVVPFLKRLSY---STPDAYGQVCQRLGWTTPRLPNTPEEAEAFVAALRDVRA-LQGPVR 70
DV LK + + S +A Q +RL RL E +A++R ++A LQG ++
Sbjct 247 DVCTKLKEVYHVDESNLEALQQESRRLMEEVERLEKEKENEPDRLASMRKLKASLQGDIQ 306
Query 71 RWLADLGESVSTSTTWSQ 88
++ L E S ST W Q
Sbjct 307 KYQNYLTEIESHSTLWDQ 324
> dre:393309 ap4b1l, MGC63878, zgc:63878; adaptor-related protein
complex 4, beta 1 subunit-like; K12401 AP-4 complex subunit
beta-1
Length=729
Score = 30.0 bits (66), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Query 146 VLAEHIKPKTKDLSTQAAALCAAAGV----FVTAWQQQQV 181
+L EH+ +D+S +LCA A V F WQQ +V
Sbjct 592 LLTEHVPVSMRDVSEAGVSLCAGAAVSPEAFEQMWQQLEV 631
> tpv:TP04_0530 DNA repair protein Rad2; K10846 DNA excision repair
protein ERCC-5
Length=835
Score = 29.3 bits (64), Expect = 8.9, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query 100 HSKVPLFL-PSSSKCHCVRSNALGRCYIDILKTTEDLIFRLSLLL 143
H VP + PS ++ C N G+CY I ++ L+F LL
Sbjct 537 HFGVPYIVAPSEAESQCAYMNRSGKCYAVISDDSDSLVFGAKCLL 581
> dre:100148910 hypothetical LOC100148910
Length=153
Score = 29.3 bits (64), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 40 WTTPRLPNTPEEAEAFVAALRDVRALQGPVRRWLADLGESVSTS 83
W T + PEE E V L+ R L G VRR+ + L + S
Sbjct 61 WYTGNIEVPPEELEDDVGLLKKGRGLAGVVRRFSSRLSSGIRNS 104
Lambda K H
0.319 0.131 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5623228644
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40