bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
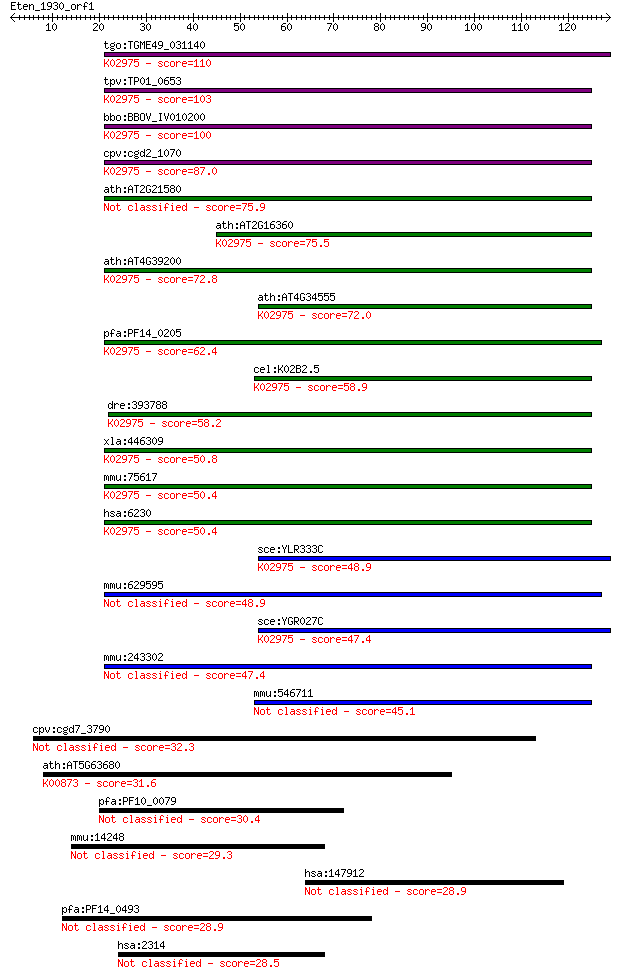
Query= Eten_1930_orf1
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_031140 R1 protein ; K02975 small subunit ribosomal ... 110 1e-24
tpv:TP01_0653 40S ribosomal protein S25; K02975 small subunit ... 103 1e-22
bbo:BBOV_IV010200 23.m06473; ribosomal protein S25; K02975 sma... 100 1e-21
cpv:cgd2_1070 40S ribosomal protein S25 ; K02975 small subunit... 87.0 1e-17
ath:AT2G21580 40S ribosomal protein S25 (RPS25B) 75.9 3e-14
ath:AT2G16360 40S ribosomal protein S25 (RPS25A); K02975 small... 75.5 4e-14
ath:AT4G39200 40S ribosomal protein S25 (RPS25E); K02975 small... 72.8 2e-13
ath:AT4G34555 40S ribosomal protein S25, putative; K02975 smal... 72.0 4e-13
pfa:PF14_0205 40S ribosomal protein S25, putative; K02975 smal... 62.4 4e-10
cel:K02B2.5 rps-25; Ribosomal Protein, Small subunit family me... 58.9 3e-09
dre:393788 rps25, MGC73391, zgc:73391; ribosomal protein S25; ... 58.2 7e-09
xla:446309 rps25, MGC82151; ribosomal protein S25; K02975 smal... 50.8 1e-06
mmu:75617 Rps25, 2810009D21Rik; ribosomal protein S25; K02975 ... 50.4 1e-06
hsa:6230 RPS25; ribosomal protein S25; K02975 small subunit ri... 50.4 1e-06
sce:YLR333C RPS25B; Rps25bp; K02975 small subunit ribosomal pr... 48.9 3e-06
mmu:629595 Gm6988, EG629595; predicted gene 6988 48.9
sce:YGR027C RPS25A, RPS31A; Rps25ap; K02975 small subunit ribo... 47.4 1e-05
mmu:243302 Gm4963, EG243302; predicted gene 4963 47.4
mmu:546711 40S ribosomal protein S25-like 45.1 5e-05
cpv:cgd7_3790 unconventional myosin 32.3 0.38
ath:AT5G63680 pyruvate kinase, putative (EC:2.7.1.40); K00873 ... 31.6 0.59
pfa:PF10_0079 conserved Plasmodium protein 30.4 1.3
mmu:14248 Flii, 3632430F08Rik, Fli1, Fliih; flightless I homol... 29.3 3.2
hsa:147912 SIX5, BOR2, DMAHP; SIX homeobox 5 28.9
pfa:PF14_0493 sortilin, putative 28.9 4.2
hsa:2314 FLII, FLI, FLIL, Fli1, MGC39265; flightless I homolog... 28.5 5.0
> tgo:TGME49_031140 R1 protein ; K02975 small subunit ribosomal
protein S25e
Length=108
Score = 110 bits (274), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 68/108 (62%), Positives = 90/108 (83%), Gaps = 0/108 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
MAPKEKKTKEQIAAAAAAGS+K+KKKWSKGKSKDKLN++V+FD T +K L+++PK +L+
Sbjct 1 MAPKEKKTKEQIAAAAAAGSRKNKKKWSKGKSKDKLNHSVLFDKATYDKLLADIPKARLI 60
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTRPAAA 128
TP ++CER ++NASLAR+ IR L Q ++ +G+ HHS YI+TR +A
Sbjct 61 TPHTVCERLKVNASLARQAIRYLKDQGLIKIVGEHHHSQYIYTRNTSA 108
> tpv:TP01_0653 40S ribosomal protein S25; K02975 small subunit
ribosomal protein S25e
Length=105
Score = 103 bits (257), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 72/104 (69%), Gaps = 0/104 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
MA KEKK+KE +A AA AG + +KKW K ++KDKLN +V+FD +K ++E+PK KL+
Sbjct 1 MAIKEKKSKEAVARAAMAGGRSKRKKWVKVRAKDKLNNSVLFDKTAYDKLMNEIPKSKLI 60
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
T + ER ++N SLAR+ +REL ++P+ + HH+ ++T+
Sbjct 61 TVSVVSERLKVNGSLARQALRELESLNLIKPICEPHHAQLLYTK 104
> bbo:BBOV_IV010200 23.m06473; ribosomal protein S25; K02975 small
subunit ribosomal protein S25e
Length=104
Score = 100 bits (249), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/104 (44%), Positives = 74/104 (71%), Gaps = 1/104 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
MA KEKK+KE + AA A S+ +KKW K K+KDK N+AV FD T +K +S++PK +L+
Sbjct 1 MAVKEKKSKEAVMRAAMA-SRGKRKKWVKVKTKDKANHAVFFDKATYDKLMSDIPKARLI 59
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
T +I ++ ++NAS+AR+ + L Q+ ++P+ DQHH+ ++T+
Sbjct 60 TISTIVDKLKVNASMARKALAHLEEQKLIKPVCDQHHAQRLYTK 103
> cpv:cgd2_1070 40S ribosomal protein S25 ; K02975 small subunit
ribosomal protein S25e
Length=106
Score = 87.0 bits (214), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 52/105 (49%), Positives = 75/105 (71%), Gaps = 1/105 (0%)
Query 21 MAPK-EKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKL 79
M PK EKKTKEQIAAAAAAG + +KKWSKGK KDK N++V+FD T ++F S+V K +L
Sbjct 1 MPPKGEKKTKEQIAAAAAAGGRAKRKKWSKGKQKDKANHSVLFDKKTADRFHSDVLKSRL 60
Query 80 VTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
+TP + + ++NAS AR +R+ + ++P+G+Q H I+T+
Sbjct 61 LTPAVVVNQLKVNASAARAMLRDCESKGIIKPVGEQTHGQMIYTK 105
> ath:AT2G21580 40S ribosomal protein S25 (RPS25B)
Length=107
Score = 75.9 bits (185), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 52/104 (50%), Positives = 67/104 (64%), Gaps = 1/104 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
MAPK+ K + A +G K KKKWSKGK K+K+N V+FD GT +K L+E PK KL+
Sbjct 1 MAPKKDKVPPPSSKPAKSGGGKQKKKWSKGKQKEKVNNMVLFDQGTYDKLLTEAPKFKLI 60
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
TP + +R RIN SLARR IREL + +R + H S I+TR
Sbjct 61 TPSILSDRMRINGSLARRAIRELMAKGLIR-MVSAHSSQQIYTR 103
> ath:AT2G16360 40S ribosomal protein S25 (RPS25A); K02975 small
subunit ribosomal protein S25e
Length=125
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 38/80 (47%), Positives = 52/80 (65%), Gaps = 1/80 (1%)
Query 45 KKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELY 104
+K SKGK K+K+N V+FD T +K +SE PK KL+TP + +R RIN SLAR+ IR+L
Sbjct 42 RKRSKGKQKEKVNNMVLFDQATYDKLMSEAPKFKLITPSILSDRLRINGSLARKAIRDLM 101
Query 105 RQQQLRPLGDQHHSHYIFTR 124
+ +R + H S I TR
Sbjct 102 VKGTIR-MVSTHSSQQINTR 120
> ath:AT4G39200 40S ribosomal protein S25 (RPS25E); K02975 small
subunit ribosomal protein S25e
Length=107
Score = 72.8 bits (177), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 51/104 (49%), Positives = 66/104 (63%), Gaps = 1/104 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
MAPK+ K + A +G K KKKWSKGK K+K+N V+FD T +K L+E PK KL+
Sbjct 1 MAPKKDKVPPPSSKPAKSGGGKQKKKWSKGKQKEKVNNMVLFDQATYDKLLTEAPKFKLI 60
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
TP + +R RIN SLARR IREL + +R + H S I+TR
Sbjct 61 TPSILSDRMRINGSLARRAIRELMAKGVIRMVA-AHSSQQIYTR 103
> ath:AT4G34555 40S ribosomal protein S25, putative; K02975 small
subunit ribosomal protein S25e
Length=108
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/71 (49%), Positives = 46/71 (64%), Gaps = 1/71 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
+K+N V+FD T +K LSE PK KL+TP + +R RIN SLARR IREL + +R +
Sbjct 35 EKVNNMVLFDQATYDKLLSEAPKFKLITPSILSDRLRINGSLARRAIRELMAKGTIR-MV 93
Query 114 DQHHSHYIFTR 124
H S I+TR
Sbjct 94 SAHSSQQIYTR 104
> pfa:PF14_0205 40S ribosomal protein S25, putative; K02975 small
subunit ribosomal protein S25e
Length=105
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 50/106 (47%), Positives = 70/106 (66%), Gaps = 1/106 (0%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLV 80
M PKE+KTKEQIAAAAAA + KKKW KGK+K+KLN+AV D K L E MK++
Sbjct 1 MPPKERKTKEQIAAAAAASGRTKKKKWGKGKNKEKLNHAVFIDKSLHSKIL-ECKNMKVI 59
Query 81 TPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTRPA 126
TP +I E++++N S+AR I L ++ + + Q HS ++T+ A
Sbjct 60 TPSAIAEKYKVNLSVARAVINHLADKKLIAEVCVQSHSQKLYTKVA 105
> cel:K02B2.5 rps-25; Ribosomal Protein, Small subunit family
member (rps-25); K02975 small subunit ribosomal protein S25e
Length=117
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 44/72 (61%), Gaps = 1/72 (1%)
Query 53 KDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPL 112
+DKLN V+FD T +K EV KL+TP + ER ++ ASLA+ G++EL + ++ +
Sbjct 37 RDKLNNMVLFDQATYDKLYKEVITYKLITPSVVSERLKVRASLAKAGLKELQAKGLVKCV 96
Query 113 GDQHHSHYIFTR 124
HH ++TR
Sbjct 97 V-HHHGQVVYTR 107
> dre:393788 rps25, MGC73391, zgc:73391; ribosomal protein S25;
K02975 small subunit ribosomal protein S25e
Length=124
Score = 58.2 bits (139), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 55/103 (53%), Gaps = 4/103 (3%)
Query 22 APKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLSEVPKMKLVT 81
A K KK K+ + + KK K +DKLN V+FD T +K EVP KL+T
Sbjct 12 AGKSKKDKDPVNKSGGKAKKKKWSKGK---VRDKLNNLVLFDKATYDKLYKEVPNYKLIT 68
Query 82 PFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
P + ER +I SLAR ++EL + ++ L +H + I+TR
Sbjct 69 PAVVSERLKIRGSLARAALQELLGKGLIK-LVSKHRAQVIYTR 110
> xla:446309 rps25, MGC82151; ribosomal protein S25; K02975 small
subunit ribosomal protein S25e
Length=125
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 9/112 (8%)
Query 21 MAPKEKKTKEQIAAAAAAG--------SKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K KKKWSKGK +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
EVP KL+TP + ER +I SLAR ++EL + ++ L +H + I+TR
Sbjct 61 EVPNYKLITPAVVSERLKIRGSLARAALQELLNKGLIK-LVSKHRAQVIYTR 111
> mmu:75617 Rps25, 2810009D21Rik; ribosomal protein S25; K02975
small subunit ribosomal protein S25e
Length=125
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 9/112 (8%)
Query 21 MAPKEKKTKEQIAAAAAAG--------SKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K KKKWSKGK +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
EVP KL+TP + ER +I SLAR ++EL + ++ L +H + I+TR
Sbjct 61 EVPNYKLITPAVVSERLKIRGSLARAALQELLSKGLIK-LVSKHRAQVIYTR 111
> hsa:6230 RPS25; ribosomal protein S25; K02975 small subunit
ribosomal protein S25e
Length=125
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 9/112 (8%)
Query 21 MAPKEKKTKEQIAAAAAAG--------SKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K KKKWSKGK +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
EVP KL+TP + ER +I SLAR ++EL + ++ L +H + I+TR
Sbjct 61 EVPNYKLITPAVVSERLKIRGSLARAALQELLSKGLIK-LVSKHRAQVIYTR 111
> sce:YLR333C RPS25B; Rps25bp; K02975 small subunit ribosomal
protein S25e
Length=108
Score = 48.9 bits (115), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 43/75 (57%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
D+ +AV+ D ++ L EVP + V+ + +R +I SLAR +R L ++ ++P+
Sbjct 34 DRAQHAVILDQEKYDRILKEVPTYRYVSVSVLVDRLKIGGSLARIALRHLEKEGIIKPIS 93
Query 114 DQHHSHYIFTRPAAA 128
+H I+TR AA+
Sbjct 94 -KHSKQAIYTRAAAS 107
> mmu:629595 Gm6988, EG629595; predicted gene 6988
Length=125
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/114 (34%), Positives = 57/114 (50%), Gaps = 9/114 (7%)
Query 21 MAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKS--------KDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K K K +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTRPA 126
EVP KL+TP + ER +I +SLAR ++EL + ++ L +H + I+ R A
Sbjct 61 EVPNYKLITPAVVSERLKIRSSLARAALQELLSKGLIK-LVSKHRAQVIYPRNA 113
> sce:YGR027C RPS25A, RPS31A; Rps25ap; K02975 small subunit ribosomal
protein S25e
Length=108
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 42/75 (56%), Gaps = 1/75 (1%)
Query 54 DKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLG 113
D+ +AV+ D ++ L EVP + V+ + +R +I SLAR +R L ++ ++P+
Sbjct 34 DRAQHAVILDQEKYDRILKEVPTYRYVSVSVLVDRLKIGGSLARIALRHLEKEGIIKPIS 93
Query 114 DQHHSHYIFTRPAAA 128
+H I+TR A+
Sbjct 94 -KHSKQAIYTRATAS 107
> mmu:243302 Gm4963, EG243302; predicted gene 4963
Length=125
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 9/112 (8%)
Query 21 MAPKEKKTKEQIAAAAAAG--------SKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFLS 72
M PK+ K K+ +A K KKKWSKGK +DKLN V+FD T +K
Sbjct 1 MPPKDDKKKKDAGKSAKKDKDPVNKSGGKAKKKKWSKGKVRDKLNNLVLFDKATYDKLCK 60
Query 73 EVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHSHYIFTR 124
EV KL+TP + ER +I +SLAR ++EL + L+ L +H + I+TR
Sbjct 61 EVQNYKLITPAVVSERLKICSSLARAALQELLSKGLLK-LVSKHRAQVIYTR 111
> mmu:546711 40S ribosomal protein S25-like
Length=127
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/72 (34%), Positives = 40/72 (55%), Gaps = 6/72 (8%)
Query 53 KDKLNYAVMFDAGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPL 112
+DKL+ V+FD EV KL+TP + ER +I SLAR +++L + ++ L
Sbjct 39 RDKLDNLVLFD-----NLCKEVSNYKLITPSMVSERLKIPGSLARAALQKLLSKGLIK-L 92
Query 113 GDQHHSHYIFTR 124
+H + I+TR
Sbjct 93 VSKHRAQMIYTR 104
> cpv:cgd7_3790 unconventional myosin
Length=816
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 29/108 (26%), Positives = 52/108 (48%), Gaps = 12/108 (11%)
Query 6 FYSFLFNSVFKNCDRMAPKEKKTKEQIAAA-AAAGSKKSKKKWSKGKSKDKLNYAVMFDA 64
F F++ FK CD A +KKT +I A +G+ K W+ GK+ ++F +
Sbjct 701 FSEFIYQ--FKYCDMSAANDKKTDPKILAEKMLSGTNIPKNSWAIGKT-------MVFLS 751
Query 65 GTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPL 112
K ++++ + KL + + + A R +R +R+ QL+PL
Sbjct 752 KDAAKRMAQLQREKLAAFEPLVQ--LLEAVYLRYKLRREFRKNQLKPL 797
> ath:AT5G63680 pyruvate kinase, putative (EC:2.7.1.40); K00873
pyruvate kinase [EC:2.7.1.40]
Length=510
Score = 31.6 bits (70), Expect = 0.59, Method: Composition-based stats.
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 14/91 (15%)
Query 8 SFLFNSVFKNCDRMAPKEKKTKEQIAAAAAAGSKKSKKK----WSKGKSKDKLNYAVMFD 63
S +N++FK R P T E +A++A + K+K K ++G + KL
Sbjct 358 SLDYNTIFKEMIRATPLPMSTLESLASSAVRTANKAKAKLIIVLTRGGTTAKL------- 410
Query 64 AGTLEKFLSEVPKMKLVTPFSICERFRINAS 94
+ K+ VP + +V P + F + S
Sbjct 411 ---VAKYRPAVPILSVVVPVFTSDTFNWSCS 438
> pfa:PF10_0079 conserved Plasmodium protein
Length=3853
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 9/52 (17%)
Query 20 RMAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFL 71
R AP E + QI +K+++K K +DKLN + MFDA L+ +L
Sbjct 2490 RKAPTEILRRYQIL---------REKEYNKNKLEDKLNISKMFDAYLLQSYL 2532
> mmu:14248 Flii, 3632430F08Rik, Fli1, Fliih; flightless I homolog
(Drosophila)
Length=1271
Score = 29.3 bits (64), Expect = 3.2, Method: Composition-based stats.
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 14 VFKNCDRMAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMF--DAGTL 67
V K +A ++ K +E+ A A G K ++W +G K +L+Y+ F D G L
Sbjct 445 VLKGMSDVAQEKNKNQEESIDARAPGGKV--RRWDQGLEKPRLDYSEFFTEDVGQL 498
> hsa:147912 SIX5, BOR2, DMAHP; SIX homeobox 5
Length=739
Score = 28.9 bits (63), Expect = 3.9, Method: Composition-based stats.
Identities = 17/55 (30%), Positives = 24/55 (43%), Gaps = 0/55 (0%)
Query 64 AGTLEKFLSEVPKMKLVTPFSICERFRINASLARRGIRELYRQQQLRPLGDQHHS 118
AG L +FL +P + + R R + R ELYR + RP HH+
Sbjct 104 AGRLSRFLGALPPAERLRGSDPVLRARALVAFQRGEYAELYRLLESRPFPAAHHA 158
> pfa:PF14_0493 sortilin, putative
Length=895
Score = 28.9 bits (63), Expect = 4.2, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 12 NSVFKNCDRMAPKEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMFDAGTLEKFL 71
N+ K+ D K E+ A ++K KK+++KGK++D + + F+ G +L
Sbjct 411 NNEIKDEDLRFQNFKSAIEEDIAPFETNTEKRKKQYNKGKNEDAVRTVISFNKGGEWSYL 470
Query 72 SEVPKM 77
+ PK+
Sbjct 471 -KAPKV 475
> hsa:2314 FLII, FLI, FLIL, Fli1, MGC39265; flightless I homolog
(Drosophila)
Length=1269
Score = 28.5 bits (62), Expect = 5.0, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query 24 KEKKTKEQIAAAAAAGSKKSKKKWSKGKSKDKLNYAVMF--DAGTL 67
+EK K++ +A A A S K ++ W +G K +L+Y+ F D G L
Sbjct 454 QEKNKKQEESADARAPSGKVRR-WDQGLEKPRLDYSEFFTEDVGQL 498
Lambda K H
0.322 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2049573556
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40