bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
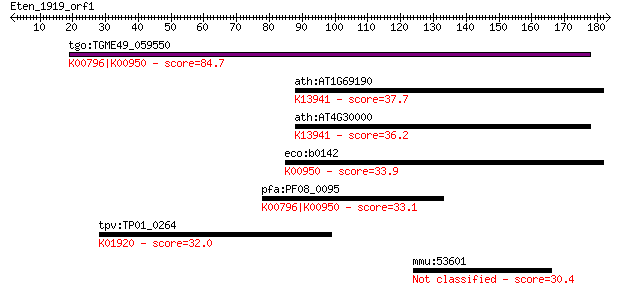
Query= Eten_1919_orf1
Length=183
Score E
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_059550 hydroxymethyldihydropterin pyrophosphokinase... 84.7 1e-16
ath:AT1G69190 2-amino-4-hydroxy-6-hydroxymethyldihydropteridin... 37.7 0.023
ath:AT4G30000 dihydropterin pyrophosphokinase, putative / dihy... 36.2 0.072
eco:b0142 folK, ECK0141, JW0138; 2-amino-4-hydroxy-6-hydroxyme... 33.9 0.29
pfa:PF08_0095 DHPS, PPPK; dihydropteroate synthetase (EC:2.5.1... 33.1 0.47
tpv:TP01_0264 glutathione synthetase; K01920 glutathione synth... 32.0 1.3
mmu:53601 Pcdh12, Pcdh14, VE-cad-2; protocadherin 12 30.4
> tgo:TGME49_059550 hydroxymethyldihydropterin pyrophosphokinase-dihydropteroate
synthase (EC:2.7.6.3 2.5.1.15); K00796 dihydropteroate
synthase [EC:2.5.1.15]; K00950 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase [EC:2.7.6.3]
Length=748
Score = 84.7 bits (208), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 53/159 (33%), Positives = 87/159 (54%), Gaps = 9/159 (5%)
Query 19 QPGSGGVILFCVHTATMFHLTSPRTSLQWFYKRCFSFVNGRSYSVRRFVRCVTTASSGES 78
+P GG I C + + F ++ F+ R F G + F++ + G+
Sbjct 38 RPLPGGAIAHCASSRS-FSFGLLQSKFSPFFTR---FHQGTMTANSHFLQKMREEDPGK- 92
Query 79 TRPKRPVAYISMGTNLGGERRLSILESALSSIRREVGSLDACSCLYESLPGYDVDHRSRD 138
+ AYI++G+NL G+ RL I+E A+S + R +G + SCLYE++P +DV +
Sbjct 93 ---EYATAYIALGSNLQGDARLGIIEEAISELGRCLGPILGTSCLYETVPAFDVCPKGV- 148
Query 139 EHDMSIPLHLNAVVRVETDSSDPQVILKILQRIEANHGR 177
HD+ P +LNAVV++ T +DP +L+IL+ +EA GR
Sbjct 149 VHDVFHPFYLNAVVKLRTPITDPWYVLEILKNLEAKTGR 187
> ath:AT1G69190 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase/ dihydropteroate synthase; K13941 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase
/ dihydropteroate synthase [EC:2.7.6.3 2.5.1.15]
Length=484
Score = 37.7 bits (86), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 13/94 (13%)
Query 88 ISMGTNLGGERRLSILESALSSIRREVGSLDACSCLYESLPGYDVDHRSRDEHDMSIPLH 147
I++G+N+G R++ + AL ++ S+ SCLYE+ P + D P
Sbjct 16 IALGSNVGN--RMNNFKEALRLMKDYGISVTRHSCLYETEPVHVTDQ----------PRF 63
Query 148 LNAVVRVETDSSDPQVILKILQRIEANHGRDRSA 181
LNA +R T P +L +L++IE GR+ +
Sbjct 64 LNAAIRGVTKLK-PHELLNVLKKIEKEMGREENG 96
> ath:AT4G30000 dihydropterin pyrophosphokinase, putative / dihydropteroate
synthase, putative / DHPS, putative; K13941 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
diphosphokinase
/ dihydropteroate synthase [EC:2.7.6.3 2.5.1.15]
Length=554
Score = 36.2 bits (82), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 13/90 (14%)
Query 88 ISMGTNLGGERRLSILESALSSIRREVGSLDACSCLYESLPGYDVDHRSRDEHDMSIPLH 147
I++G+N+G R++ AL ++R + SCLYE+ P + D P
Sbjct 91 IALGSNIGN--RMNNFREALRLMKRGGICVTRHSCLYETAPVHVTDQ----------PRF 138
Query 148 LNAVVRVETDSSDPQVILKILQRIEANHGR 177
LNA VR T P +L +L+ IE + GR
Sbjct 139 LNAAVRGVTKLG-PHELLSVLKTIERDMGR 167
> eco:b0142 folK, ECK0141, JW0138; 2-amino-4-hydroxy-6-hydroxymethyldihyropteridine
pyrophosphokinase (EC:2.7.6.3); K00950
2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase
[EC:2.7.6.3]
Length=159
Score = 33.9 bits (76), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 46/98 (46%), Gaps = 14/98 (14%)
Query 85 VAYISMGTNLGGERRLSILESALSSIRREVGS-LDACSCLYESLPGYDVDHRSRDEHDMS 143
VAYI++G+NL L + +AL ++ S + S Y + P D
Sbjct 3 VAYIAIGSNLASP--LEQVNAALKALGDIPESHILTVSSFYRTPPLGPQDQ--------- 51
Query 144 IPLHLNAVVRVETDSSDPQVILKILQRIEANHGRDRSA 181
P +LNA V +ET S P+ +L QRIE GR R A
Sbjct 52 -PDYLNAAVALET-SLAPEELLNHTQRIELQQGRVRKA 87
> pfa:PF08_0095 DHPS, PPPK; dihydropteroate synthetase (EC:2.5.1.15);
K00796 dihydropteroate synthase [EC:2.5.1.15]; K00950
2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase
[EC:2.7.6.3]
Length=706
Score = 33.1 bits (74), Expect = 0.47, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 34/57 (59%), Gaps = 5/57 (8%)
Query 78 STRPKRPVAYISMGTNLGGERR--LSILESALSSIRREVGSLDACSCLYESLPGYDV 132
S K +A +++GTN +RR + ILE+AL + + +G + S LYE++P Y V
Sbjct 10 SEENKTNIAVLNLGTN---DRRNAVLILETALHLVEKYLGKIINTSYLYETVPEYIV 63
> tpv:TP01_0264 glutathione synthetase; K01920 glutathione synthase
[EC:6.3.2.3]
Length=629
Score = 32.0 bits (71), Expect = 1.3, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 31/71 (43%), Gaps = 6/71 (8%)
Query 28 FCVHTATMFHLTSPRTSLQWFYKRCFSFVNGRSYSVRRFVRCVTTASSGESTRPKRPVAY 87
+ VH T+ +PR +QW KR F V Y+ V+ + A S P R V
Sbjct 328 YGVHVKTV----TPRQMIQWMKKRIFLLVRDWEYNSDGTVKLLKYADSTVKLHPGRLV-- 381
Query 88 ISMGTNLGGER 98
I MG+ G R
Sbjct 382 IIMGSKPGLNR 392
> mmu:53601 Pcdh12, Pcdh14, VE-cad-2; protocadherin 12
Length=1180
Score = 30.4 bits (67), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 124 YESLPGYDVDHRSRDEHDMSIPLHLNAVVRV-ETDSSDPQVIL 165
YE P Y+VD ++RD SIP H +++V + + + P +++
Sbjct 312 YEKNPAYEVDVQARDLGPNSIPGHCKVLIKVLDVNDNAPSILI 354
Lambda K H
0.322 0.134 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4976880524
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40