bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1915_orf2
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
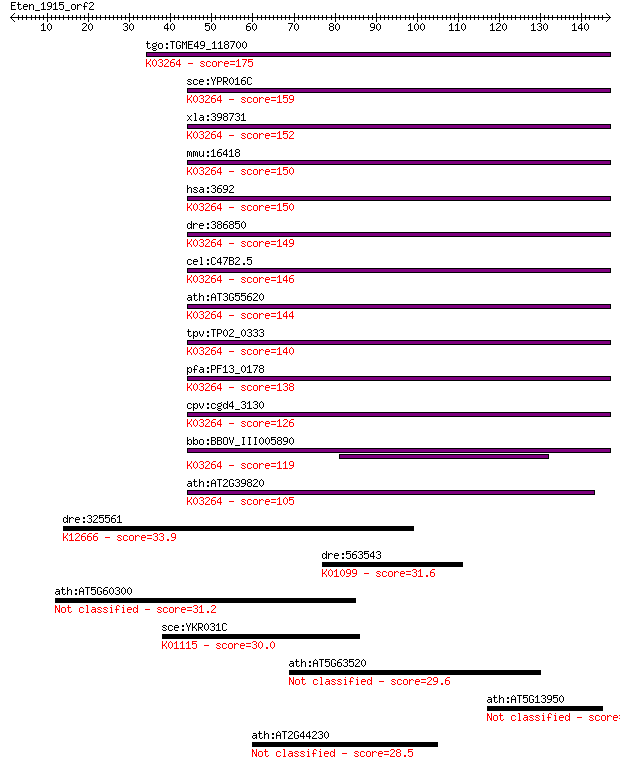
tgo:TGME49_118700 eukaryotic translation initiation factor 6, ... 175 6e-44
sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal part... 159 3e-39
xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic ... 152 5e-37
mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc... 150 1e-36
hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP), ... 150 2e-36
dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a0... 149 2e-36
cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member ... 146 2e-35
ath:AT3G55620 emb1624 (embryo defective 1624); ribosome bindin... 144 1e-34
tpv:TP02_0333 translation initiation factor 6; K03264 translat... 140 1e-33
pfa:PF13_0178 translation initiation factor 6, putative; K0326... 138 7e-33
cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264 t... 126 2e-29
bbo:BBOV_III005890 17.m07522; translation initiation factor eI... 119 2e-27
ath:AT2G39820 eukaryotic translation initiation factor 6, puta... 105 4e-23
dre:325561 rpn1, wu:fa12h07, wu:fc68b07, wu:fc88a12; ribophori... 33.9 0.21
dre:563543 inositol polyphosphate 5-phosphatase-like; K01099 p... 31.6 1.0
ath:AT5G60300 lectin protein kinase family protein 31.2 1.2
sce:YKR031C SPO14, PLD1; Phospholipase D, catalyzes the hydrol... 30.0 2.9
ath:AT5G63520 hypothetical protein 29.6 3.8
ath:AT5G13950 hypothetical protein 28.9 6.5
ath:AT2G44230 hypothetical protein 28.5 8.4
> tgo:TGME49_118700 eukaryotic translation initiation factor 6,
putative ; K03264 translation initiation factor 6
Length=264
Score = 175 bits (443), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 85/113 (75%), Positives = 93/113 (82%), Gaps = 3/113 (2%)
Query 34 WAAFSSLVNKMAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVH 93
W+ SS +MA R FESSNEVGVF KLTNSYCLVA GGSEHFYS +EAEL PHIPVVH
Sbjct 9 WSLGSS---RMATRAQFESSNEVGVFAKLTNSYCLVALGGSEHFYSTLEAELAPHIPVVH 65
Query 94 ASVAGTAVIGRVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
A+V GT VIGRVCVGN+ GL+VPS T+D ELQ LRNSLPD V +RRVEERLSA
Sbjct 66 ATVGGTRVIGRVCVGNRRGLIVPSITTDQELQHLRNSLPDSVEIRRVEERLSA 118
> sce:YPR016C TIF6, CDC95; Constituent of 66S pre-ribosomal particles,
has similarity to human translation initiation factor
6 (eIF6); may be involved in the biogenesis and or stability
of 60S ribosomal subunits; K03264 translation initiation
factor 6
Length=245
Score = 159 bits (401), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 72/103 (69%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA RT FE+SNE+GVF KLTN+YCLVA GGSE+FYSA EAELG IP+VH ++AGT +IG
Sbjct 1 MATRTQFENSNEIGVFSKLTNTYCLVAVGGSENFYSAFEAELGDAIPIVHTTIAGTRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ GN+ GLLVP+ T+D ELQ LRNSLPD V ++RVEERLSA
Sbjct 61 RMTAGNRRGLLVPTQTTDQELQHLRNSLPDSVKIQRVEERLSA 103
> xla:398731 eif6, eIF-6, itgb4bp, p27(BBP), p27BBP; eukaryotic
translation initiation factor 6 (EC:3.6.5.3); K03264 translation
initiation factor 6
Length=245
Score = 152 bits (383), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 69/103 (66%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE++NE+G F KLTN+YCLVA GGSE+FYS E EL IPVVHAS+AG +IG
Sbjct 1 MAVRASFENNNEIGCFAKLTNTYCLVAIGGSENFYSVFEGELSETIPVVHASIAGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+CVGN+HGL+VP++T+D ELQ +RNSLPD V ++RVEERLSA
Sbjct 61 RMCVGNRHGLMVPNNTTDQELQHMRNSLPDSVRIQRVEERLSA 103
> mmu:16418 Eif6, 1110004P11, AA408895, CAB, Itgb4bp, eIF-6, imc-415,
p27(BBP), p27BBP; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 150 bits (379), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 71/103 (68%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE++ EVG F KLTN+YCLVA GGSE+FYS E EL IPVVHAS+AG +IG
Sbjct 1 MAVRASFENNCEVGCFAKLTNAYCLVAIGGSENFYSVFEGELSDAIPVVHASIAGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+CVGN+HGLLVP++T+D ELQ +RNSLPD V +RRVEERLSA
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNSLPDSVQIRRVEERLSA 103
> hsa:3692 EIF6, CAB, EIF3A, ITGB4BP, b(2)gcn, eIF-6, p27(BBP),
p27BBP; eukaryotic translation initiation factor 6 (EC:3.6.5.3);
K03264 translation initiation factor 6
Length=245
Score = 150 bits (378), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE++ E+G F KLTN+YCLVA GGSE+FYS E EL IPVVHAS+AG +IG
Sbjct 1 MAVRASFENNCEIGCFAKLTNTYCLVAIGGSENFYSVFEGELSDTIPVVHASIAGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+CVGN+HGLLVP++T+D ELQ +RNSLPD V +RRVEERLSA
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNSLPDTVQIRRVEERLSA 103
> dre:386850 eif6, Itgb4bp, MGC56562, eIF-6, itgb4bp4, wu:fc28a04,
wu:ft88f05, zgc:56562; eukaryotic translation initiation
factor 6 (EC:3.6.5.3); K03264 translation initiation factor
6
Length=245
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 68/103 (66%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R FE +NE+G F KLTN+YCLVA GGSE+FYSA E EL +PV+HAS+AG +IG
Sbjct 1 MAVRASFEKNNEIGCFAKLTNTYCLVAIGGSENFYSAFEGELSETMPVIHASIAGCRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+CVGN+HGLLVP++T+D ELQ +RN LPD V ++RVEERLSA
Sbjct 61 RMCVGNRHGLLVPNNTTDQELQHIRNCLPDSVRIQRVEERLSA 103
> cel:C47B2.5 eif-6; Eukaryotic Initiation Factor family member
(eif-6); K03264 translation initiation factor 6
Length=246
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 67/103 (65%), Positives = 85/103 (82%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R +E SN+VGVF LTNSYCLV GG+++FYS +EAEL IPVVH S+A T ++G
Sbjct 1 MALRVDYEGSNDVGVFCTLTNSYCLVGVGGTQNFYSILEAELSDLIPVVHTSIASTRIVG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+ VGN+HGLLVP++T+D ELQ LRNSLPD V++RRV+ERLSA
Sbjct 61 RLTVGNRHGLLVPNATTDQELQHLRNSLPDEVAIRRVDERLSA 103
> ath:AT3G55620 emb1624 (embryo defective 1624); ribosome binding
/ translation initiation factor; K03264 translation initiation
factor 6
Length=245
Score = 144 bits (362), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 68/103 (66%), Positives = 83/103 (80%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA R FE++ EVGVF KLTN+YCLVA GGSE+FYSA E+EL IP+V S+ GT +IG
Sbjct 1 MATRLQFENNCEVGVFSKLTNAYCLVAIGGSENFYSAFESELADVIPIVKTSIGGTRIIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R+C GNK+GLLVP +T+D ELQ LRNSLPD V V+R++ERLSA
Sbjct 61 RLCAGNKNGLLVPHTTTDQELQHLRNSLPDQVVVQRIDERLSA 103
> tpv:TP02_0333 translation initiation factor 6; K03264 translation
initiation factor 6
Length=249
Score = 140 bits (353), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 68/103 (66%), Positives = 79/103 (76%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MAIR +E+SNEVGVF LTNSY LV+ G S +F S EAEL PHIPVVH ++ GT VIG
Sbjct 1 MAIRAQYENSNEVGVFSTLTNSYALVSLGSSTNFSSLFEAELTPHIPVVHTTIGGTRVIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
RV VGNK GLLV S +D EL+ LRNSLPD V +RR++ERLSA
Sbjct 61 RVSVGNKKGLLVSSICTDKELRHLRNSLPDSVEIRRIDERLSA 103
> pfa:PF13_0178 translation initiation factor 6, putative; K03264
translation initiation factor 6
Length=247
Score = 138 bits (347), Expect = 7e-33, Method: Compositional matrix adjust.
Identities = 66/103 (64%), Positives = 80/103 (77%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MAIR FE+SNEVGVF +LTNSY LVA GGSE+F S E+EL HIP+V+ ++ GT VIG
Sbjct 1 MAIRVQFENSNEVGVFSRLTNSYALVALGGSENFSSVFESELSQHIPLVYTTIGGTRVIG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
RVCVGN+ GLLV S +D EL LRN LP+ V ++R+EERLSA
Sbjct 61 RVCVGNRKGLLVSSICTDQELLHLRNCLPENVKIKRIEERLSA 103
> cpv:cgd4_3130 eIF6, translation initiation factor 6 ; K03264
translation initiation factor 6
Length=252
Score = 126 bits (317), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 59/103 (57%), Positives = 76/103 (73%), Gaps = 0/103 (0%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MA+R +ESS+E+GVF LTN YCL+A G S F S EAEL HIP+++ + GT ++G
Sbjct 1 MALRVAYESSSEIGVFANLTNRYCLLAHGSSAAFTSVFEAELMDHIPIINTLIGGTRLVG 60
Query 104 RVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
R VGN++GLLV + +D ELQ LRNSLPD V V+R+EERLSA
Sbjct 61 RCTVGNRNGLLVSNMATDQELQHLRNSLPDNVKVQRIEERLSA 103
> bbo:BBOV_III005890 17.m07522; translation initiation factor
eIF-6 family protein; K03264 translation initiation factor 6
Length=262
Score = 119 bits (299), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 63/116 (54%), Positives = 77/116 (66%), Gaps = 13/116 (11%)
Query 44 MAIRTHFESSNEVGVFGKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIG 103
MAIRT +E+SNEVGVF LTNSY LV+ G S +F S EAEL P IPVV ++ GT V+G
Sbjct 1 MAIRTQYENSNEVGVFATLTNSYALVSLGSSCNFASVFEAELMPQIPVVQTTIGGTRVVG 60
Query 104 RVCV-------------GNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEERLSA 146
V V GN+ GLLV S +D EL+ LRNSLPD V +RR+++RLSA
Sbjct 61 SVTVGMLYSNYSVILFTGNRKGLLVSSICTDTELRHLRNSLPDSVEIRRIDDRLSA 116
Score = 28.9 bits (63), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query 81 IEAELGPHIPVVHASVAGTAVIGRVCVGNKHGLLVPSSTSDGELQQLRNSL 131
+E LG I V AS+AG +IG C G LV T+ E+++L L
Sbjct 142 VEDVLG--IEVFRASIAGNVLIGSYCRFQNKGGLVHVKTTTDEMEELSQLL 190
> ath:AT2G39820 eukaryotic translation initiation factor 6, putative
/ eIF-6, putative; K03264 translation initiation factor
6
Length=247
Score = 105 bits (263), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/101 (50%), Positives = 74/101 (73%), Gaps = 2/101 (1%)
Query 44 MAIRTHFESSN-EVGVFGKLTNSYCLV-ASGGSEHFYSAIEAELGPHIPVVHASVAGTAV 101
MA R ++++N E+GVF KLTN+YCLV A+ S +F++ E++L IP+V S+ G+
Sbjct 1 MATRLQYDNNNCEIGVFSKLTNAYCLVSATSASANFFTGYESKLKGVIPIVTTSIGGSGT 60
Query 102 IGRVCVGNKHGLLVPSSTSDGELQQLRNSLPDGVSVRRVEE 142
IG +CVGNK+GLL+ + +D ELQ LR+SLPD V V+R+EE
Sbjct 61 IGSLCVGNKNGLLLSHTITDQELQHLRDSLPDEVVVQRIEE 101
> dre:325561 rpn1, wu:fa12h07, wu:fc68b07, wu:fc88a12; ribophorin
I (EC:2.4.1.119); K12666 oligosaccharyltransferase complex
subunit alpha (ribophorin I)
Length=598
Score = 33.9 bits (76), Expect = 0.21, Method: Composition-based stats.
Identities = 31/89 (34%), Positives = 39/89 (43%), Gaps = 14/89 (15%)
Query 14 RSATVTFGPI--FLCGIIFRPGWAAFSSLVNKMAIRTHFESSNEVGVFGKLTNSYCLVAS 71
R+ V+F I F CG + G LVN+ RT SS+ K+T L
Sbjct 3 RAGAVSFALICCFFCGSVCADG------LVNEDVKRTLDLSSH----LAKITAEIQLANR 52
Query 72 GGSE--HFYSAIEAELGPHIPVVHASVAG 98
G S F +E EL PH+ V ASV G
Sbjct 53 GSSRANSFIIGLEEELAPHLAFVGASVKG 81
> dre:563543 inositol polyphosphate 5-phosphatase-like; K01099
phosphatidylinositol-bisphosphatase [EC:3.1.3.36]
Length=677
Score = 31.6 bits (70), Expect = 1.0, Method: Composition-based stats.
Identities = 17/34 (50%), Positives = 21/34 (61%), Gaps = 4/34 (11%)
Query 77 FYSAIEAELGPHIPVVHASVAGTAVIGRVCVGNK 110
FY ++AE PHI V A GT V+GR +GNK
Sbjct 234 FY--VKAEHAPHISEVEAETVGTGVMGR--MGNK 263
> ath:AT5G60300 lectin protein kinase family protein
Length=766
Score = 31.2 bits (69), Expect = 1.2, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 36/76 (47%), Gaps = 10/76 (13%)
Query 12 FLRSATVTFGPIFLCGIIFRPGWAAFSSLVNKMAIR---THFESSNEVGVFGKLTNSYCL 68
F S ++F F+C ++ +PG+ +V ++ TH ES+ +G+F TN
Sbjct 73 FSSSGPLSFSTHFVCALVPKPGFEGGHGIVFVLSPSMDFTHAESTRYLGIFNASTN---- 128
Query 69 VASGGSEHFYSAIEAE 84
G S + A+E +
Sbjct 129 ---GSSSYHVLAVELD 141
> sce:YKR031C SPO14, PLD1; Phospholipase D, catalyzes the hydrolysis
of phosphatidylcholine, producing choline and phosphatidic
acid; involved in Sec14p-independent secretion; required
for meiosis and spore formation; differently regulated in
secretion and meiosis (EC:3.1.4.4); K01115 phospholipase D
[EC:3.1.4.4]
Length=1683
Score = 30.0 bits (66), Expect = 2.9, Method: Composition-based stats.
Identities = 18/49 (36%), Positives = 27/49 (55%), Gaps = 2/49 (4%)
Query 38 SSLVNKMAIRTHFESSNEVGVFGKL-TNSYCLVASGGSEHFYSAIEAEL 85
SS++ KM+ T + N G F + TNS+C G ++F+S EA L
Sbjct 655 SSII-KMSTSTPWSKPNRFGSFAPVRTNSFCKFLVDGRDYFWSLSEALL 702
> ath:AT5G63520 hypothetical protein
Length=519
Score = 29.6 bits (65), Expect = 3.8, Method: Composition-based stats.
Identities = 17/61 (27%), Positives = 30/61 (49%), Gaps = 2/61 (3%)
Query 69 VASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIGRVCVGNKHGLLVPSSTSDGELQQLR 128
+ G E + I +G +P++ + V G ++G+ +K G + STSD EL +
Sbjct 101 ITCGNMEETLTLITERVGSRVPIIVSVVTG--ILGKEACNDKAGEVRLHSTSDDELFDVA 158
Query 129 N 129
N
Sbjct 159 N 159
> ath:AT5G13950 hypothetical protein
Length=939
Score = 28.9 bits (63), Expect = 6.5, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 117 SSTSDGELQQLRNSLPDGVSVRRVEERL 144
S SDGE LR LP+GV V +V + L
Sbjct 110 SCLSDGERNYLRQFLPEGVDVEQVVQAL 137
> ath:AT2G44230 hypothetical protein
Length=542
Score = 28.5 bits (62), Expect = 8.4, Method: Composition-based stats.
Identities = 18/45 (40%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 60 GKLTNSYCLVASGGSEHFYSAIEAELGPHIPVVHASVAGTAVIGR 104
GKL Y SGGS S IE + G + PV +AS+ G A+ +
Sbjct 385 GKLHRMYLSQHSGGSWADASEIEFQGGGNKPVAYASLNGHAMYSK 429
Lambda K H
0.325 0.138 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2872883024
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40