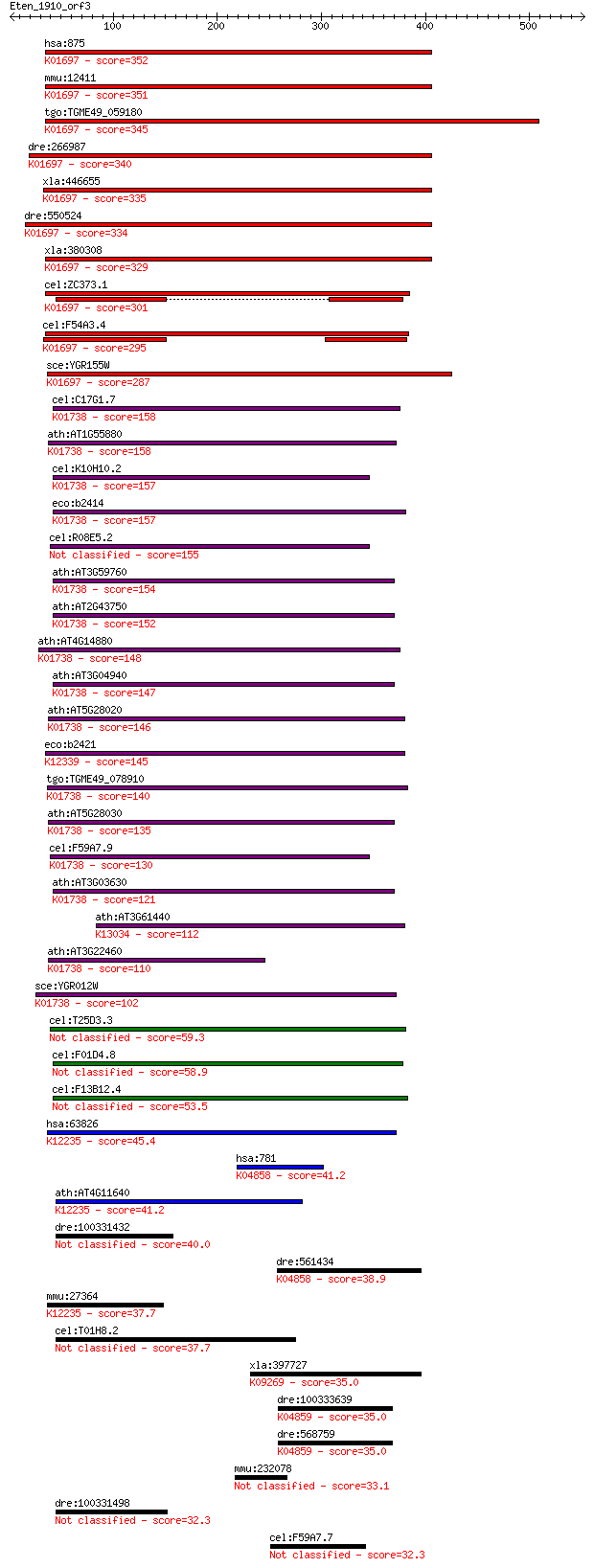
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1910_orf3
Length=552
Score E
Sequences producing significant alignments: (Bits) Value
hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22); ... 352 3e-96
mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895, M... 351 5e-96
tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.... 345 3e-94
dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c0... 340 7e-93
xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synt... 335 3e-91
dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-syn... 334 5e-91
xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase ... 329 2e-89
cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-sy... 301 6e-81
cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-sy... 295 2e-79
sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K016... 287 8e-77
cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A [... 158 5e-38
ath:AT1G55880 pyridoxal-5'-phosphate-dependent enzyme, beta fa... 158 5e-38
cel:K10H10.2 hypothetical protein; K01738 cysteine synthase A ... 157 7e-38
eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A, O-... 157 1e-37
cel:R08E5.2 hypothetical protein 155 5e-37
ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM... 154 7e-37
ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cys... 152 3e-36
ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-... 148 5e-35
ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine sy... 147 1e-34
ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/ ... 146 2e-34
eco:b2421 cysM, ECK2416, JW2414; cysteine synthase B (O-acetyl... 145 4e-34
tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2... 140 2e-32
ath:AT5G28030 cysteine synthase, putative / O-acetylserine (th... 135 4e-31
cel:F59A7.9 hypothetical protein; K01738 cysteine synthase A [... 130 1e-29
ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine s... 121 7e-27
ath:AT3G61440 CYSC1; CYSC1 (CYSTEINE SYNTHASE C1); L-3-cyanoal... 112 5e-24
ath:AT3G22460 OASA2; OASA2 (O-ACETYLSERINE (THIOL) LYASE (OAS-... 110 1e-23
sce:YGR012W Putative cysteine synthase, localized to the mitoc... 102 3e-21
cel:T25D3.3 hypothetical protein 59.3 3e-08
cel:F01D4.8 hypothetical protein 58.9 5e-08
cel:F13B12.4 hypothetical protein 53.5 2e-06
hsa:63826 SRR, ILV1, ISO1; serine racemase (EC:4.3.1.17 5.1.1.... 45.4 6e-04
hsa:781 CACNA2D1, CACNA2, CACNL2A, CCHL2A; calcium channel, vo... 41.2 0.010
ath:AT4G11640 ATSR; ATSR (ARABIDOPSIS THALIANA SERINE RACEMASE... 41.2 0.010
dre:100331432 serine racemase-like 40.0 0.021
dre:561434 cacna2d1a, si:ch211-251k4.1; calcium channel, volta... 38.9 0.060
mmu:27364 Srr, Srs; serine racemase (EC:4.3.1.17 5.1.1.18 4.3.... 37.7 0.11
cel:T01H8.2 hypothetical protein 37.7 0.13
xla:397727 sox13, Sry, sox12, xSox12, xsox13; SRY (sex determi... 35.0 0.75
dre:100333639 calcium channel, voltage-dependent, alpha 2/delt... 35.0 0.80
dre:568759 novel protein similar to vertebrate calcium channel... 35.0 0.80
mmu:232078 Thnsl2, BC051244, MGC59076, TSH2; threonine synthas... 33.1 3.3
dre:100331498 serine racemase-like 32.3 4.4
cel:F59A7.7 hypothetical protein 32.3 5.2
> hsa:875 CBS, HIP4; cystathionine-beta-synthase (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=551
Score = 352 bits (902), Expect = 3e-96, Method: Compositional matrix adjust.
Identities = 176/374 (47%), Positives = 239/374 (63%), Gaps = 34/374 (9%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
LP IL IG TP+V ++++G G+ C+LL KCEF +AGGS+KDRI+ MI AE G L
Sbjct 77 LPDILKKIGDTPMVRINKIGKKFGLKCELLAKCEFFNAGGSVKDRISLRMIEDAERDGTL 136
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
PG +IEPTSGNTGIG+AL AAV+GYR + VMP+KMS+EK ++RALG+ I+RTPT A
Sbjct 137 KPGDTIIEPTSGNTGIGLALAAAVRGYRCIIVMPEKMSSEKVDVLRALGAEIVRTPTNAR 196
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
+D SH+ ++ RLK E+ ILDQYRN +NP
Sbjct 197 FDSPESHVGVAWRLKNEIPNSH---------------------------ILDQYRNASNP 229
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
++H+ T E+L QC +LDM+V GTGG++TGI R++K P C ++ VDPEGSILA
Sbjct 230 LAHYDTTADEILQQCDGKLDMLVASVGTGGTITGIARKLKEKCPGCRIIGVDPEGSILAE 289
Query 275 P---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGS 331
P N+ Y VEGIGYDF+PTVLDR V D W K ND E+ +R+LI EG+L GGS
Sbjct 290 PEELNQTEQTTYEVEGIGYDFIPTVLDRTVVDKWFKSNDEEAFTFARMLIAQEGLLCGGS 349
Query 332 SGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQ 391
+G+ +A A+KA + + +R ++LPD RNY +KF+ D WM +KGFL+ +L+ +
Sbjct 350 AGSTVAVAVKAAQEL----QEGQRCVVILPDSVRNYMTKFLSDRWMLQKGFLKEEDLTEK 405
Query 392 YEQYSNMSVQSLQL 405
+ ++ VQ L L
Sbjct 406 KPWWWHLRVQELGL 419
> mmu:12411 Cbs, AI047524, AI303044, HIP4, MGC18856, MGC18895,
MGC37300; cystathionine beta-synthase (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=561
Score = 351 bits (900), Expect = 5e-96, Method: Compositional matrix adjust.
Identities = 181/374 (48%), Positives = 239/374 (63%), Gaps = 35/374 (9%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
LP IL IG TP+V ++++ AG+ C+LL KCEF +AGGS+KDRI+ MI AE +G L
Sbjct 74 LPDILRKIGNTPMVRINKISKNAGLKCELLAKCEFFNAGGSVKDRISLRMIEDAERAGNL 133
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK ++RALG+ I+RTPT A
Sbjct 134 KPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTNAR 193
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
+D SH+ ++ RLK E+ ILDQYRN +NP
Sbjct 194 FDSPESHVGVAWRLKNEIPNSH---------------------------ILDQYRNASNP 226
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
++H+ T E+L QC +LDM+V GTGG++TGI R++K P C ++ VDPEGSILA
Sbjct 227 LAHYDDTAEEILQQCDGKLDMLVASAGTGGTITGIARKLKEKCPGCKIIGVDPEGSILAE 286
Query 275 P---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGS 331
P N+ Y VEGIGYDF+PTVLDR V D W K ND +S +R+LI EG+L GGS
Sbjct 287 PEELNQTEQTAYEVEGIGYDFIPTVLDRAVVDKWFKSNDEDSFAFARMLIAQEGLLCGGS 346
Query 332 SGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQ 391
SG+A+A A+KA + +R ++LPD RNY SKF+ D+WM +KGF+ + ELS +
Sbjct 347 SGSAMAVAVKAAREL----QEGQRCVVILPDSVRNYMSKFLSDKWMLQKGFM-KEELSVK 401
Query 392 YEQYSNMSVQSLQL 405
+ + VQ L L
Sbjct 402 RPWWWRLRVQELSL 415
> tgo:TGME49_059180 cystathionine beta-synthase, putative (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=514
Score = 345 bits (884), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 204/487 (41%), Positives = 292/487 (59%), Gaps = 52/487 (10%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
L S+LDAIG TP+V + R+ G+ C LL KCEF+SAGGS+KDRI + M+ AE GRL
Sbjct 14 LDSVLDAIGNTPMVRMKRLAKVYGLECDLLAKCEFMSAGGSVKDRIGKAMVEKAEREGRL 73
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
G LIEPTSGNTGIG+AL AAV+GYR + MP KMSAEK +IM+ LG+ I+RTPT+AA
Sbjct 74 KAGDTLIEPTSGNTGIGLALAAAVRGYRMIVTMPAKMSAEKSNIMKCLGAEIVRTPTEAA 133
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
W+D+ SH+ ++ +L+ E+E A ILDQY N ANP
Sbjct 134 WNDENSHMGVAAKLQRELEN---------------------------AHILDQYNNTANP 166
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
+ H+ T E++ QC +DMVVI GTGG++TGIGR+IK P C VV VDP+GSILA
Sbjct 167 MVHYDVTAEEIITQCDGDIDMVVIGAGTGGTITGIGRKIKERCPKCKVVGVDPKGSILAV 226
Query 275 P----NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGG 330
P +E+ + Y VEGIGYDFVP VLDR V D WVKV DAES +R +IR EG+ VGG
Sbjct 227 PDSLNDEKRLQSYEVEGIGYDFVPGVLDRKVVDEWVKVGDAESFTTARAIIRNEGLFVGG 286
Query 331 SSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFL--ERSEL 388
SSGA + GAL+A ++ ++ ++LPD SRNY SKF+ DEWMAE GF + +++
Sbjct 287 SSGANVWGALQAARQL----KKGQKCVVLLPDSSRNYMSKFISDEWMAEHGFAPEDGAKV 342
Query 389 SRQYEQYSNMSVQSL-----QLPRLTFVSSDSSLKEAAALLVQSPQPILAVIEPGAAEDA 443
+ +Q+ ++ L + FV++ S+++ ++ ++ + V E A+
Sbjct 343 KEREKQFGGARIRDLLSETGATSDVPFVTARLSVEDVIKMMHETKVKEVIVTEDSEADGK 402
Query 444 TQNGTAPRQMVTGSVTQKALLMAL--GEMPSSAPIGEIADTETPVYNIGTPLSLLAQSLE 501
T+ + G +++ + +L G +P+ +IA + L +A++L+
Sbjct 403 TK--------LVGVLSEDHIAHSLQSGRCAMQSPVKDIAFKKLAKALPSAYLRDVAKALD 454
Query 502 LHPFLFI 508
P++ +
Sbjct 455 FSPYVCV 461
> dre:266987 cbsb, cb442, cbs, wu:fb37g05, wu:fm61c07, wu:fq06c06,
zgc:113704; cystathionine-beta-synthase b (EC:4.2.1.22);
K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=597
Score = 340 bits (873), Expect = 7e-93, Method: Compositional matrix adjust.
Identities = 184/390 (47%), Positives = 250/390 (64%), Gaps = 34/390 (8%)
Query 19 ISSPPAAFFQSHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKD 78
+ SP + ++ + LP+IL IG TP+V ++++ A G+ C+LL KCEF +AGGS+KD
Sbjct 96 MESPHNHYQRTKTSGILPNILSKIGETPMVRMNKIPKAFGLKCELLAKCEFFNAGGSVKD 155
Query 79 RIARDMIVTAEASGRLAPGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSI 138
RI+ MI AE G L PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK +
Sbjct 156 RISLRMIEDAERDGTLKPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDV 215
Query 139 MRALGSMILRTPTKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDE 198
+RALG+ I+RTPT A +D SH+ ++ RLK E+ P+
Sbjct 216 LRALGAEIVRTPTSARFDSPESHVGVAWRLKNEI-----------------------PN- 251
Query 199 LQIACILDQYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALP 258
A ILDQYRNP+NP++H+ T E+L QCG ++DM+V GTGG++TGI R++K P
Sbjct 252 ---AHILDQYRNPSNPLAHYDTTAEEILEQCGGKIDMLVAGAGTGGTITGIARKLKEKCP 308
Query 259 NCLVVAVDPEGSILANP---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLL 315
+ ++ VDPEGSILA P N+ Y VEGIGYDF+PTVLDR V D W K ND ES
Sbjct 309 DIKIIGVDPEGSILAEPDELNKTDKTQYEVEGIGYDFIPTVLDRSVVDNWYKSNDEESFA 368
Query 316 CSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDE 375
SR+LIR EG+L GGSSG A++ A+ + + +R ++LPD RNY SKF+ D+
Sbjct 369 MSRMLIREEGLLCGGSSGTAMSAAVNVAKEL----KEGQRCVVILPDSIRNYMSKFLSDK 424
Query 376 WMAEKGFLERSELSRQYEQYSNMSVQSLQL 405
WM EKGF+ +L + N+++Q L+L
Sbjct 425 WMFEKGFMSEEDLVVNKPWWWNLTLQELRL 454
> xla:446655 cbs-b, MGC82640, cbs, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=562
Score = 335 bits (859), Expect = 3e-91, Method: Compositional matrix adjust.
Identities = 183/376 (48%), Positives = 239/376 (63%), Gaps = 34/376 (9%)
Query 33 RELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASG 92
R L IL IG TPLV ++++G G+ C+LL KCEF +AGGS+KDRI+ M+ AE G
Sbjct 68 RILSCILQKIGDTPLVRINQIGRRFGLKCELLAKCEFFNAGGSVKDRISLRMVEDAERDG 127
Query 93 RLAPGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTK 152
L PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK ++RALG+ I+RTPT
Sbjct 128 ILKPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTS 187
Query 153 AAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPA 212
A +D SH+ ++ RLK E+ ILDQYRN +
Sbjct 188 ARFDSPESHVGVAWRLKNEIPNSH---------------------------ILDQYRNAS 220
Query 213 NPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSIL 272
NP++H+ T E+L QC QLDM+V GTGG++TG+ R++K PNC +V VDPEGSIL
Sbjct 221 NPLAHYDSTAEEILQQCEGQLDMLVAGAGTGGTITGLARKLKEKCPNCKIVGVDPEGSIL 280
Query 273 ANP---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVG 329
A P N+ Y VEGIGYDF+PTVLDR V D W K ND ES +R LIR EG+L G
Sbjct 281 AEPEELNQTDKTGYEVEGIGYDFIPTVLDRTVVDEWYKSNDEESFFMARALIREEGLLCG 340
Query 330 GSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELS 389
GSSG+A++ A+KA + + +R ++LPD RNY SKF+ D+WM +K FL+ +L
Sbjct 341 GSSGSAMSVAVKAAKDLS----EGQRCVVILPDSVRNYMSKFLSDKWMTQKQFLKVEKLE 396
Query 390 RQYEQYSNMSVQSLQL 405
+ + N+ V SL L
Sbjct 397 ERKPWWWNLKVSSLSL 412
> dre:550524 cbsa, MGC174292, zgc:110199; cystathionine-beta-synthase
a; K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=571
Score = 334 bits (857), Expect = 5e-91, Method: Compositional matrix adjust.
Identities = 183/393 (46%), Positives = 246/393 (62%), Gaps = 34/393 (8%)
Query 16 LKYISSPPAAFFQSHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGS 75
+ + +SP F ++ LP+IL IG TPLV ++++ G+ C+LL KCEF +AGGS
Sbjct 69 MSHENSPHTYFPRTKAECILPNILGKIGDTPLVHINKISKIFGLKCELLAKCEFFNAGGS 128
Query 76 MKDRIARDMIVTAEASGRLAPGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEK 135
+KDRI+ M+ AE G L PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK
Sbjct 129 VKDRISLRMVEDAEREGILNPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSMEK 188
Query 136 QSIMRALGSMILRTPTKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKET 195
++RALG+ I+RTPT A +D SH+ ++ RLK E+
Sbjct 189 VDVLRALGAEIVRTPTSARFDSPESHVGVAWRLKNEIPNSH------------------- 229
Query 196 PDELQIACILDQYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKL 255
ILDQYRNP+NP++H+ T E+L QC ++DM+V GTGG++TGI R++K
Sbjct 230 --------ILDQYRNPSNPLAHYDTTAEEILEQCDGKVDMLVAGAGTGGTITGIARKLKE 281
Query 256 ALPNCLVVAVDPEGSILANPNE---QPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAE 312
PN +VAVDPEGSILA P+E Y VEGIGYDF+PTVLDR + D W K D E
Sbjct 282 KCPNIKIVAVDPEGSILAEPDELNRTDKNQYEVEGIGYDFIPTVLDRSLVDKWYKSKDEE 341
Query 313 SLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFV 372
S +R+LIR EG+L GGSSG A+A A+K + + +R ++LPD RNY SKF+
Sbjct 342 SFAMARMLIRDEGLLCGGSSGTAMAAAVKFAKEL----KEGQRCVVILPDSIRNYMSKFL 397
Query 373 CDEWMAEKGFLERSELSRQYEQYSNMSVQSLQL 405
D+WM +KGFL ++ + N+ +QSL L
Sbjct 398 SDKWMFQKGFLRVEDIMVNKPWWWNLKLQSLNL 430
> xla:380308 cbs-a, MGC52737, hip4; cystathionine-beta-synthase
(EC:4.2.1.22); K01697 cystathionine beta-synthase [EC:4.2.1.22]
Length=565
Score = 329 bits (843), Expect = 2e-89, Method: Compositional matrix adjust.
Identities = 184/374 (49%), Positives = 240/374 (64%), Gaps = 34/374 (9%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
L SIL IG TPLV ++++G G+ C+LL KCEF +AGGS+KDRI+ M+ AE G L
Sbjct 73 LSSILQKIGDTPLVRINQIGKRFGLKCELLAKCEFFNAGGSVKDRISLRMVEDAERDGIL 132
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
PG +IEPTSGNTGIG+AL AAVKGYR + VMP+KMS EK ++RALG+ I+RTPT A
Sbjct 133 KPGDTIIEPTSGNTGIGLALAAAVKGYRCIIVMPEKMSMEKVDVLRALGAEIVRTPTSAR 192
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
+D SH+ ++ RLK E+ ILDQYRN +NP
Sbjct 193 FDSPESHVGVAWRLKNEIPNSH---------------------------ILDQYRNASNP 225
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
++H+ T E+L QC QLDM+V GTGG++TG+ R++K PNC +V VDP+GSILA
Sbjct 226 LAHYDSTAEEILQQCQGQLDMLVAGAGTGGTITGLARKLKEKCPNCKIVGVDPDGSILAE 285
Query 275 P---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGS 331
P N+ Y VEGIGYDF+PTVLDR V D W K ND ES +R LIR EG+L GGS
Sbjct 286 PEELNQTDKTGYEVEGIGYDFIPTVLDRTVVDEWYKSNDEESFFMARALIREEGLLCGGS 345
Query 332 SGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLERSELSRQ 391
SG+A++ A+KA + +G +R ++LPD RNY SKF+ D+WM +K FL+ EL +
Sbjct 346 SGSAMSAAMKAAKELG----EGQRCVVILPDSVRNYMSKFLSDKWMTQKQFLKVEELEER 401
Query 392 YEQYSNMSVQSLQL 405
+ N+ V SL L
Sbjct 402 KPWWWNLKVSSLSL 415
> cel:ZC373.1 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=704
Score = 301 bits (770), Expect = 6e-81, Method: Compositional matrix adjust.
Identities = 164/354 (46%), Positives = 223/354 (62%), Gaps = 33/354 (9%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGR- 93
L ++LDAIG TPLV L + A GV C + KCE+++AGGS KDRIA+ M+ AE +G+
Sbjct 379 LDTVLDAIGKTPLVKLQHIPKAHGVKCNVYVKCEYMNAGGSTKDRIAKRMVEIAEKTGKP 438
Query 94 --LAPGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPT 151
L PG LIEPTSGNTGIG++L +AV+GY+ + MPKKMS EK M +LGS I+RTP
Sbjct 439 GKLVPGVTLIEPTSGNTGIGLSLASAVRGYKCIITMPKKMSKEKSIAMASLGSTIIRTPN 498
Query 152 KAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNP 211
+A +D SHI +++RLK E +Q A +LDQY NP
Sbjct 499 EAGFDSPHSHIGVALRLKSE---------------------------IQDAVVLDQYCNP 531
Query 212 ANPISHFFGTGPELLHQCG-MQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGS 270
NP++H+ T E+++ G +D+VV+ GTGG++TGI R+I +P VV VDP GS
Sbjct 532 GNPLAHYEETAEEIIYDMGDKHIDLVVLTAGTGGTVTGISRKIHERIPTAKVVGVDPHGS 591
Query 271 ILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGG 330
ILA P E Y VEGIGYDF+P LD D+W K +D ES L +R LIR EG+L GG
Sbjct 592 ILAGPAETDIDFYEVEGIGYDFLPGTLDTSAIDYWAKSHDKESFLMARELIRSEGILCGG 651
Query 331 SSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLE 384
SSG A+ AL+ + + +++ + ++LPDG RNY +KF+ D+WM E+ FL+
Sbjct 652 SSGCAVHYALEECKSLNLPAEAN--VVVLLPDGIRNYITKFLDDDWMNERHFLD 703
Score = 46.2 bits (108), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 4/71 (5%)
Query 307 KVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRN 366
+V + E+ +R LI EG++ G SSGAA+ A+K + + + I +VL DG RN
Sbjct 250 EVEEDEAYSFTRHLIGTEGIMAGPSSGAAVLEAIKLAKEL----PAGSTIVVVLMDGIRN 305
Query 367 YTSKFVCDEWM 377
Y F+ D+W+
Sbjct 306 YLRHFLDDDWI 316
Score = 44.7 bits (104), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/106 (29%), Positives = 47/106 (44%), Gaps = 1/106 (0%)
Query 45 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALIEPT 104
TPLV L ++ + + K E+L+ GS++DR A AE G + G +
Sbjct 30 TPLVPLKKLKVEHQLQSDIYVKLEYLNIAGSLEDRTADKAFQFAEEIG-VVRGDEVFVTA 88
Query 105 SGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTP 150
G+ I A VAAVKG + PK +++ LG + P
Sbjct 89 GGSAAISYATVAAVKGIKLTIYAPKGEFDLVDTVLHTLGVKTVELP 134
> cel:F54A3.4 hypothetical protein; K01697 cystathionine beta-synthase
[EC:4.2.1.22]
Length=755
Score = 295 bits (756), Expect = 2e-79, Method: Compositional matrix adjust.
Identities = 169/355 (47%), Positives = 225/355 (63%), Gaps = 35/355 (9%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGR- 93
L S+LDAIG TPLV L V A GV C + KCEFL+AGGS KDRIA+ M+ AE +G+
Sbjct 337 LDSVLDAIGKTPLVKLQHVPKAHGVRCNVYVKCEFLNAGGSTKDRIAKKMVEIAEKTGKP 396
Query 94 --LAPGAA-LIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTP 150
L PGA LIEPTSGNTGIG++LVAAV+GY+ + MP+KMS EK + + LGS I+RTP
Sbjct 397 GALTPGATTLIEPTSGNTGIGLSLVAAVRGYKCLITMPEKMSKEKSTTLSVLGSTIVRTP 456
Query 151 TKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRN 210
+AA++ +SHI +++RLK E+ A ILDQY N
Sbjct 457 NEAAFNSPSSHIGVALRLKHEIPG---------------------------AVILDQYCN 489
Query 211 PANPISHFFGTGPELLHQCG-MQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEG 269
P NP++H+ T E+L G ++D+VV+ GTGG++TGI R+I PN +VV VDP G
Sbjct 490 PGNPLAHYEETAEEILWDMGDRKIDLVVLGAGTGGTITGISRKIHERRPNAIVVGVDPNG 549
Query 270 SILANPNEQPGKP-YLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLV 328
SIL P P Y VEGIGYDF+P LD D W+K +D ES L +R +IR EG+L
Sbjct 550 SILTGPTTGPAPDFYEVEGIGYDFIPGTLDEKSVDSWLKSDDKESFLMAREIIRTEGILC 609
Query 329 GGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFL 383
GGSSG A+ AL+ ++ +D++ + ++LPDG RNY +KF+ D+WM +GF
Sbjct 610 GGSSGCAVHYALEQCRKLDLPEDAN--VVVLLPDGIRNYLTKFLDDDWMKARGFF 662
Score = 44.7 bits (104), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 56/120 (46%), Gaps = 5/120 (4%)
Query 33 RELPSILDAIG--CTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEA 90
+ +ILDA T LV L ++ + + K E+L+ GS++DR A E
Sbjct 7 KAFKTILDAGSHRATQLVPLKKLKAEFQLKSDIYVKLEYLNISGSLEDRAAHRAF---EM 63
Query 91 SGRLAPGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTP 150
S + G + G++ + A VAAVKG R A P + ++I+ LG+ I+ P
Sbjct 64 SNGIQRGDEVYVTAGGSSAVSYATVAAVKGVRLTAHAPPGAFDQVRTILSTLGTKIVELP 123
Score = 38.1 bits (87), Expect = 0.081, Method: Compositional matrix adjust.
Identities = 27/79 (34%), Positives = 42/79 (53%), Gaps = 9/79 (11%)
Query 304 FWVKVNDA-ESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPD 362
+ V++ DA ES +R LI EG++ G SSGAA+ A+K + + + + +VL D
Sbjct 227 YKVEIRDAPESYTFTRHLIETEGIMAGPSSGAAVLEAIKLAKDL----PAGSVVVVVLMD 282
Query 363 GSRNYTSKFVCDEWMAEKG 381
G R+Y +WM G
Sbjct 283 GIRDYLDA----DWMKVNG 297
> sce:YGR155W CYS4, NHS5, STR4, VMA41; Cys4p (EC:4.2.1.22); K01697
cystathionine beta-synthase [EC:4.2.1.22]
Length=507
Score = 287 bits (734), Expect = 8e-77, Method: Compositional matrix adjust.
Identities = 166/411 (40%), Positives = 230/411 (55%), Gaps = 53/411 (12%)
Query 37 SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAP 96
+++D +G TPL+ L ++ A G+ Q+ K E + GGS+KDRIA+ M+ AEASGR+ P
Sbjct 13 NVIDLVGNTPLIALKKLPKALGIKPQIYAKLELYNPGGSIKDRIAKSMVEEAEASGRIHP 72
Query 97 G-AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAW 155
+ LIEPTSGNTGIG+AL+ A+KGYR++ +P+KMS EK S+++ALG+ I+RTPT AAW
Sbjct 73 SRSTLIEPTSGNTGIGLALIGAIKGYRTIITLPEKMSNEKVSVLKALGAEIIRTPTAAAW 132
Query 156 DDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPI 215
D SHI ++ +L EKE P A ILDQY N NP
Sbjct 133 DSPESHIGVAKKL-----------------------EKEIPG----AVILDQYNNMMNPE 165
Query 216 SHFFGTGPELLHQCGM-----QLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGS 270
+H+FGTG E+ Q L VV GTGG+++GI + +K +V DP GS
Sbjct 166 AHYFGTGREIQRQLEDLNLFDNLRAVVAGAGTGGTISGISKYLKEQNDKIQIVGADPFGS 225
Query 271 ILANP---NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGML 327
ILA P N+ Y VEGIGYDFVP VLDR + D W K +D S +R LI EG+L
Sbjct 226 ILAQPENLNKTDITDYKVEGIGYDFVPQVLDRKLIDVWYKTDDKPSFKYARQLISNEGVL 285
Query 328 VGGSSGAALAGALKAIERMG--WKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGFLER 385
VGGSSG+A +K E +DD I + PD R+Y +KFV DEW+ + +
Sbjct 286 VGGSSGSAFTAVVKYCEDHPELTEDDV---IVAIFPDSIRSYLTKFVDDEWLKKNNLWDD 342
Query 386 SELSR-----------QY-EQYSNMSVQSLQLPRLTFVSSDSSLKEAAALL 424
L+R +Y + + N +V+ L L + V + + + +L
Sbjct 343 DVLARFDSSKLEASTTKYADVFGNATVKDLHLKPVVSVKETAKVTDVIKIL 393
> cel:C17G1.7 hypothetical protein; K01738 cysteine synthase A
[EC:2.5.1.47]
Length=341
Score = 158 bits (400), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 119/336 (35%), Positives = 180/336 (53%), Gaps = 41/336 (12%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAA-L 100
IG TP+V ++++ G+ + K E+++ GS+KDRI M+ AE G + PG L
Sbjct 15 IGNTPMVYINKL--TKGLPGTVAVKIEYMNPAGSVKDRIGAAMLAAAEKDGTVIPGVTTL 72
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGS-MILRTPTKAAWDDQT 159
IEPTSGNTGI +A VAA KGYR + MP MS E++++++A GS ++L P K
Sbjct 73 IEPTSGNTGIALAFVAAAKGYRCIVTMPASMSGERRTLLKAYGSEVVLTDPAKG----MK 128
Query 160 SHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFF 219
I ++ +LKE + P + IL Q+ NP NP+ H+
Sbjct 129 GAIDMANQLKENI-----------------------PGSI----ILAQFDNPNNPLVHYQ 161
Query 220 GTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILANPNEQP 279
TGPE+ Q +D VV GTGG++TG+GR ++ P V AV+PE S + + +P
Sbjct 162 TTGPEIWRQTKGTVDAVVFGVGTGGTITGVGRYLQEQNPGVRVFAVEPEESAIL--SGRP 219
Query 280 GKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGA 339
P+ ++GIG F P VLD + + ++++ E+++ ++ L EG+L G SSGA +A A
Sbjct 220 AGPHKIQGIGAGFAPAVLDTKIYEDVIRIHSDEAIVMAQRLSYEEGLLGGISSGANVAAA 279
Query 340 LKAIERMGWKDDSSKRIAMVLPD-GSRNYTSKFVCD 374
L+ R + + K I LP G R TS D
Sbjct 280 LQLAAR---PEMAGKLIVTCLPSCGERYMTSPLYTD 312
> ath:AT1G55880 pyridoxal-5'-phosphate-dependent enzyme, beta
family protein (EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=421
Score = 158 bits (400), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 116/361 (32%), Positives = 174/361 (48%), Gaps = 34/361 (9%)
Query 38 ILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG 97
++DAIG TPL+ ++ + A G C++LGKCEFL+ GGS+KDR+A +I A SG+L PG
Sbjct 44 LVDAIGNTPLIRINSLSEATG--CEILGKCEFLNPGGSVKDRVAVKIIQEALESGKLFPG 101
Query 98 AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRT-PTKAAWD 156
+ E ++G+T I +A VA G + V+P + EK I+ ALG+ + R P
Sbjct 102 GIVTEGSAGSTAISLATVAPAYGCKCHVVIPDDAAIEKSQIIEALGASVERVRPVSITHK 161
Query 157 DQTSHIALSIRLKEEMERQRKEK--------KKDDSNDCYETPEKE---TPDELQIACIL 205
D +IA R E E K + ++ +N C KE D +
Sbjct 162 DHYVNIARR-RADEANELASKRRLGSETNGIHQEKTNGCTVEEVKEPSLFSDSVTGGFFA 220
Query 206 DQYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAV 265
DQ+ N AN +H+ GTGPE+ HQ +D V GTGG+L G+ R ++ +
Sbjct 221 DQFENLANYRAHYEGTGPEIWHQTQGNIDAFVAAAGTGGTLAGVSRFLQDKNERVKCFLI 280
Query 266 DPEGSILANP---------NEQPGK----PY--LVEGIGYDFVPTVLDRDVADFWVKVND 310
DP GS L N E G+ P+ + EGIG + + D + D
Sbjct 281 DPPGSGLYNKVTRGVMYTREEAEGRRLKNPFDTITEGIGINRLTKNFLMAKLDGGFRGTD 340
Query 311 AESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSK 370
E++ SR L++ +G+ VG SS GA++ + +G I +L D + SK
Sbjct 341 KEAVEMSRFLLKNDGLFVGSSSAMNCVGAVRVAQTLG----PGHTIVTILCDSGMRHLSK 396
Query 371 F 371
F
Sbjct 397 F 397
> cel:K10H10.2 hypothetical protein; K01738 cysteine synthase
A [EC:2.5.1.47]
Length=337
Score = 157 bits (398), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 100/306 (32%), Positives = 168/306 (54%), Gaps = 37/306 (12%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-AAL 100
IG TPL+ L+++G G S + K E+++ S+KDRIA +MI TAE +G + PG L
Sbjct 14 IGNTPLLKLNKIGKDLGASIAV--KVEYMNPACSVKDRIAFNMIDTAEKAGLITPGKTVL 71
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTS 160
IEPTSGN GI +A ++GY+ + MP MS E++ +++A G+ ++ T D +
Sbjct 72 IEPTSGNMGIALAYCGKLRGYKVILTMPASMSIERRCLLKAYGAEVILT-------DPAT 124
Query 161 HIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFG 220
+ +++ EE+ D + A IL+Q+ NPANP +H+
Sbjct 125 AVKGAVQRAEELR-----------------------DVIPNAYILNQFGNPANPEAHYKT 161
Query 221 TGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNEQP 279
TGPE+ Q ++D+V G+GG+ TG+GR +K P+ V V+P E S++ N P
Sbjct 162 TGPEIWRQTQGKVDIVCFGVGSGGTCTGVGRFLKEKNPSVQVFPVEPFESSVI---NGLP 218
Query 280 GKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGA 339
P+ ++G+G +P +LD + ++V+ +++ ++ L E +L G SSGA + A
Sbjct 219 HSPHKIQGMGTGMIPDILDLTLFSEALRVHSDDAIAMAKKLADEESILGGISSGANVCAA 278
Query 340 LKAIER 345
++ +R
Sbjct 279 VQLAKR 284
> eco:b2414 cysK, cysZ, ECK2409, JW2407; cysteine synthase A,
O-acetylserine sulfhydrolase A subunit (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 157 bits (396), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 122/346 (35%), Positives = 180/346 (52%), Gaps = 45/346 (13%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALI 101
IG TPLV L+R+G ++L K E + S+K RI +MI AE G L PG L+
Sbjct 12 IGHTPLVRLNRIGNG-----RILAKVESRNPSFSVKCRIGANMIWDAEKRGVLKPGVELV 66
Query 102 EPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTSH 161
EPTSGNTGI +A VAA +GY+ MP+ MS E++ +++ALG+ ++ T +
Sbjct 67 EPTSGNTGIALAYVAAARGYKLTLTMPETMSIERRKLLKALGANLVLT-------EGAKG 119
Query 162 IALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFGT 221
+ +I+ EE+ PEK +L Q+ NPANP H T
Sbjct 120 MKGAIQKAEEI--------------VASNPEK--------YLLLQQFSNPANPEIHEKTT 157
Query 222 GPELLHQCGMQLDMVVICTGTGGSLTGIGRRIK--LALPNCLVVAVDPEGS-----ILAN 274
GPE+ Q+D+ + GTGG+LTG+ R IK + + VAV+P S LA
Sbjct 158 GPEIWEDTDGQVDVFIAGVGTGGTLTGVSRYIKGTKGKTDLISVAVEPTDSPVIAQALAG 217
Query 275 PNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGA 334
+PG P+ ++GIG F+P LD + D + + + E++ +R L+ EG+L G SSGA
Sbjct 218 EEIKPG-PHKIQGIGAGFIPANLDLKLVDKVIGITNEEAISTARRLMEEEGILAGISSGA 276
Query 335 ALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEK 380
A+A ALK E + ++K I ++LP Y S + + EK
Sbjct 277 AVAAALKLQEDESF---TNKNIVVILPSSGERYLSTALFADLFTEK 319
> cel:R08E5.2 hypothetical protein
Length=337
Score = 155 bits (391), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 102/309 (33%), Positives = 164/309 (53%), Gaps = 39/309 (12%)
Query 40 DAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-A 98
D IG TPLV L + + G+ ++ K E+L+ S+KDRIA+ M+ AE +G + PG
Sbjct 12 DTIGNTPLVLLRNI--SKGLDARIAVKVEYLNPSCSVKDRIAKSMVDEAEKAGTIVPGKT 69
Query 99 ALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGS-MILRTPTKAAWDD 157
L+E TSGN GI +A + ++GY+ + VMP MS E+++++RA G+ +IL P +
Sbjct 70 VLVEGTSGNLGIALAHIGKIRGYKVILVMPATMSVERRAMLRAYGAEVILSDPAEGHP-- 127
Query 158 QTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISH 217
+ K EM D+L A LDQ+ NPANP +H
Sbjct 128 -------GVIKKVEM----------------------LVDKLPNAHCLDQFSNPANPAAH 158
Query 218 FFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPN 276
+ TGPE+ Q ++D+V G+ G++TG+GR ++ PN + V+P E S+L+
Sbjct 159 YRTTGPEIWRQTEGKVDIVCFGVGSSGTVTGVGRYLREQNPNIEIYPVEPYESSVLSG-- 216
Query 277 EQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAAL 336
P P+ ++GIG VP +DR + +++ +++ +R L E +L G SSGA +
Sbjct 217 -LPRGPHKIQGIGAGIVPGNVDRSLFTEILRIKSDDAMQMARRLADEEAILGGISSGANV 275
Query 337 AGALKAIER 345
A++ R
Sbjct 276 VAAVELASR 284
> ath:AT3G59760 OASC; OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM
C); ATP binding / cysteine synthase (EC:2.5.1.47); K01738
cysteine synthase A [EC:2.5.1.47]
Length=430
Score = 154 bits (390), Expect = 7e-37, Method: Compositional matrix adjust.
Identities = 118/330 (35%), Positives = 181/330 (54%), Gaps = 40/330 (12%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-AAL 100
IG TP+V L+ + A G + K E + S+KDRI M+ AE G ++PG + L
Sbjct 121 IGKTPMVYLNSI--AKGCVANIAAKLEIMEPCCSVKDRIGYSMVTDAEQKGFISPGKSVL 178
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTS 160
+EPTSGNTGIG+A +AA +GYR + MP MS E++ +++A G+ ++ T D
Sbjct 179 VEPTSGNTGIGLAFIAASRGYRLILTMPASMSMERRVLLKAFGAELVLT-------DPAK 231
Query 161 HIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFG 220
+ +++ EE+ K TPD A +L Q+ NPANP H+
Sbjct 232 GMTGAVQKAEEI-------------------LKNTPD----AYMLQQFDNPANPKIHYET 268
Query 221 TGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNEQP 279
TGPE+ ++D+ V GTGG++TG+GR IK P V+ V+P E IL+ +P
Sbjct 269 TGPEIWDDTKGKVDIFVAGIGTGGTITGVGRFIKEKNPKTQVIGVEPTESDILS--GGKP 326
Query 280 GKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGA 339
G P+ ++GIG F+P LD+ + D + ++ E++ ++ L EG++VG SSGAA A A
Sbjct 327 G-PHKIQGIGAGFIPKNLDQKIMDEVIAISSEEAIETAKQLALKEGLMVGISSGAAAAAA 385
Query 340 LKAIERMGWKDDSSKRIAMVLPDGSRNYTS 369
+K +R +++ K IA+V P Y S
Sbjct 386 IKVAKR---PENAGKLIAVVFPSFGERYLS 412
> ath:AT2G43750 OASB; OASB (O-ACETYLSERINE (THIOL) LYASE B); cysteine
synthase; K01738 cysteine synthase A [EC:2.5.1.47]
Length=392
Score = 152 bits (385), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 121/331 (36%), Positives = 181/331 (54%), Gaps = 42/331 (12%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-AAL 100
IG TP+V L+ V G + K E + S+KDRI MI AE G + PG + L
Sbjct 83 IGKTPMVYLNNV--VKGCVASVAAKLEIMEPCCSVKDRIGYSMITDAEEKGLITPGKSVL 140
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGS-MILRTPTKAAWDDQT 159
+E TSGNTGIG+A +AA KGY+ + MP MS E++ ++RA G+ ++L P K
Sbjct 141 VESTSGNTGIGLAFIAASKGYKLILTMPASMSLERRVLLRAFGAELVLTEPAKG------ 194
Query 160 SHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFF 219
+ +I+ EE+ K+TP+ +L Q+ NPANP H+
Sbjct 195 --MTGAIQKAEEIL-------------------KKTPNSY----MLQQFDNPANPKIHYE 229
Query 220 GTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNEQ 278
TGPE+ ++D++V GTGG++TG+GR IK P V+ V+P E +IL+ +
Sbjct 230 TTGPEIWEDTRGKIDILVAGIGTGGTITGVGRFIKERKPELKVIGVEPTESAILS--GGK 287
Query 279 PGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAG 338
PG P+ ++GIG FVP LD + D ++ ++ E++ S+ L EG+LVG SSGAA A
Sbjct 288 PG-PHKIQGIGAGFVPKNLDLAIVDEYIAISSEEAIETSKQLALQEGLLVGISSGAAAAA 346
Query 339 ALKAIERMGWKDDSSKRIAMVLPDGSRNYTS 369
A++ +R +++ K IA+V P Y S
Sbjct 347 AIQVAKR---PENAGKLIAVVFPSFGERYLS 374
> ath:AT4G14880 OASA1; OASA1 (O-ACETYLSERINE (THIOL) LYASE (OAS-TL)
ISOFORM A1); cysteine synthase (EC:2.5.1.47); K01738 cysteine
synthase A [EC:2.5.1.47]
Length=322
Score = 148 bits (374), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 125/349 (35%), Positives = 185/349 (53%), Gaps = 45/349 (12%)
Query 29 SHLARELPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTA 88
S +A++ + + IG TPLV L+ V A G ++ K E + S+KDRI MI A
Sbjct 3 SRIAKD---VTELIGNTPLVYLNNV--AEGCVGRVAAKLEMMEPCSSVKDRIGFSMISDA 57
Query 89 EASGRLAPG-AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALG-SMI 146
E G + PG + LIEPTSGNTG+G+A AA KGY+ + MP MS E++ I+ A G ++
Sbjct 58 EKKGLIKPGESVLIEPTSGNTGVGLAFTAAAKGYKLIITMPASMSTERRIILLAFGVELV 117
Query 147 LRTPTKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILD 206
L P K + +I EE+ + TP+ +L
Sbjct 118 LTDPAKG--------MKGAIAKAEEILAK-------------------TPN----GYMLQ 146
Query 207 QYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVD 266
Q+ NPANP H+ TGPE+ G ++D V GTGG++TG G+ +K N + V+
Sbjct 147 QFENPANPKIHYETTGPEIWKGTGGKIDGFVSGIGTGGTITGAGKYLKEQNANVKLYGVE 206
Query 267 P-EGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEG 325
P E +IL+ +PG P+ ++GIG F+P+VL+ D+ D V+V+ ES+ +R L EG
Sbjct 207 PVESAILSG--GKPG-PHKIQGIGAGFIPSVLNVDLIDEVVQVSSDESIDMARQLALKEG 263
Query 326 MLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCD 374
+LVG SSGAA A A+K +R +++ K + P Y S + D
Sbjct 264 LLVGISSGAAAAAAIKLAQR---PENAGKLFVAIFPSFGERYLSTVLFD 309
> ath:AT3G04940 CYSD1; CYSD1 (CYSTEINE SYNTHASE D1); cysteine
synthase (EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=324
Score = 147 bits (370), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 116/332 (34%), Positives = 180/332 (54%), Gaps = 44/332 (13%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-AAL 100
IG TP+V L+ + G ++ K E + S+K+RIA MI AE G + PG + L
Sbjct 16 IGNTPMVYLNNI--VDGCVARIAAKLEMMEPCSSVKERIAYGMIKDAEDKGLITPGKSTL 73
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTS 160
IE TSGNTGIG+A + A KGY+ V MP MS E++ I+ ALG+ + T D +
Sbjct 74 IEATSGNTGIGLAFIGAAKGYKVVLTMPSSMSLERKIILLALGAEVHLT-------DPSK 126
Query 161 HIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFG 220
+ I EE+ C + P+ + +L+Q++NP+NP +H+
Sbjct 127 GVQGIIDKAEEI--------------CSKNPD---------SIMLEQFKNPSNPQTHYRT 163
Query 221 TGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNEQP 279
TGPE+ ++D++V GTGG+L+G GR +K + V V+P E ++++
Sbjct 164 TGPEIWRDSAGEVDILVAGVGTGGTLSGSGRFLKEKNKDFKVYGVEPTESAVISG----- 218
Query 280 GKP--YLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALA 337
GKP +L++GIG +P LD +V D ++V E++ ++LL EG+LVG SSGAA A
Sbjct 219 GKPGTHLIQGIGAGLIPDNLDFNVLDEVIQVTSVEAIETAKLLALKEGLLVGISSGAAAA 278
Query 338 GALKAIERMGWKDDSSKRIAMVLPDGSRNYTS 369
A+K +R +++ K I ++ P G Y S
Sbjct 279 AAIKVAKR---PENAGKLIVVIFPSGGERYLS 307
> ath:AT5G28020 CYSD2; CYSD2 (CYSTEINE SYNTHASE D2); catalytic/
cysteine synthase/ pyridoxal phosphate binding (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 146 bits (369), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 127/344 (36%), Positives = 181/344 (52%), Gaps = 40/344 (11%)
Query 38 ILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG 97
I + IG TP+V L+ V G ++ K E + S+KDRIA MI AE G + PG
Sbjct 11 ITELIGNTPMVYLNNV--VDGCVARIAAKLEMMEPCSSVKDRIAYSMIKDAEDKGLITPG 68
Query 98 -AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWD 156
+ LIEPT+GNTGIG+A + A +GY+ + VMP MS E++ I+RALG+ +
Sbjct 69 KSTLIEPTAGNTGIGLACMGAARGYKVILVMPSTMSLERRIILRALGA-------ELHLS 121
Query 157 DQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPIS 216
DQ I LK +E+ K TP P Q+ NPANP
Sbjct 122 DQ------RIGLKGMLEKTEAILSK--------TPGGYIP---------QQFENPANPEI 158
Query 217 HFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANP 275
H+ TGPE+ ++D++V GTGG+ TG+G+ +K + V V+P E +L+
Sbjct 159 HYRTTGPEIWRDSAGKVDILVAGVGTGGTATGVGKFLKEQNKDIKVCVVEPVESPVLS-- 216
Query 276 NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAA 335
QPG P+L++GIG VP LD + D ++V E++ ++LL EG+LVG SSGAA
Sbjct 217 GGQPG-PHLIQGIGSGIVPFNLDLTIVDEIIQVAGEEAIETAKLLALKEGLLVGISSGAA 275
Query 336 LAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAE 379
A ALK +R +++ K I +V P G Y S + D E
Sbjct 276 AAAALKVAKR---PENAGKLIVVVFPSGGERYLSTKLFDSIRYE 316
> eco:b2421 cysM, ECK2416, JW2414; cysteine synthase B (O-acetylserine
sulfhydrolase B) (EC:2.5.1.47); K12339 cysteine synthase
B [EC:2.5.1.47]
Length=303
Score = 145 bits (366), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 107/346 (30%), Positives = 167/346 (48%), Gaps = 48/346 (13%)
Query 35 LPSILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL 94
+ ++ IG TPLV L R+G G L K E + GS+KDR A MIV AE G +
Sbjct 1 MSTLEQTIGNTPLVKLQRMGPDNGSEVWL--KLEGNNPAGSVKDRAALSMIVEAEKRGEI 58
Query 95 APGAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAA 154
PG LIE TSGNTGI +A++AA+KGYR +MP MS E+++ MRA G+ ++ TK
Sbjct 59 KPGDVLIEATSGNTGIALAMIAALKGYRMKLLMPDNMSQERRAAMRAYGAELILV-TKEQ 117
Query 155 WDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANP 214
+ +AL + R E K +LDQ+ NP NP
Sbjct 118 GMEGARDLALEM-------ANRGEGK-----------------------LLDQFNNPDNP 147
Query 215 ISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILA 273
+H+ TGPE+ Q G ++ V GT G++TG+ R ++ +V + P EGS +
Sbjct 148 YAHYTTTGPEIWQQTGGRITHFVSSMGTTGTITGVSRFMREQSKPVTIVGLQPEEGSSIP 207
Query 274 NPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSG 333
P +++P + + + D + ++ ++ R L EG+ G SSG
Sbjct 208 GIRRWP----------TEYLPGIFNASLVDEVLDIHQRDAENTMRELAVREGIFCGVSSG 257
Query 334 AALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAE 379
A+AGAL R+ + + +A++ G R ++ +E ++
Sbjct 258 GAVAGAL----RVAKANPDAVVVAIICDRGDRYLSTGVFGEEHFSQ 299
> tgo:TGME49_078910 O-acetylserine (thiol) lyase, putative (EC:2.5.1.47);
K01738 cysteine synthase A [EC:2.5.1.47]
Length=499
Score = 140 bits (352), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 103/351 (29%), Positives = 158/351 (45%), Gaps = 51/351 (14%)
Query 37 SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAP 96
+ LDA+G TPL+ L R C + KCEF + GGS+KDR A+ ++ AE GRL
Sbjct 147 NFLDAVGMTPLIYLRRASQE--THCDIFAKCEFNNPGGSVKDRPAKYILEQAEKEGRLGG 204
Query 97 GAA-LIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAW 155
++E ++GNT IG+AL + +GYR V M ++ G++++ P W
Sbjct 205 SQNWIVEGSAGNTAIGLALASKPRGYRVVCDM-----------LKVCGALVVEVPFHKVW 253
Query 156 DDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPI 215
+A S RL M AC +Q+ N AN +
Sbjct 254 HPD-HFVAFSGRLASVMN----------------------------ACWGNQFDNLANQL 284
Query 216 SHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILANP 275
SH+ TGPE+ Q ++D V GTGG+++G+ R ++ P + D GS L
Sbjct 285 SHYETTGPEIWDQMDGRIDGFVSAAGTGGTVSGVSRYLRSQHPEIQIALADIPGSALYRY 344
Query 276 NEQ----PGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGS 331
+ +VE IG + L D +++D ESL + L+ EG+ VG S
Sbjct 345 FTEGVLRAEGASVVENIGQGRITGQLQDFRPDVAFEISDVESLTWTHSLLEDEGLAVGMS 404
Query 332 SGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAEKGF 382
SG +A A++ +R+G RI +L D Y K ++ E+G
Sbjct 405 SGVNVAAAVRLAKRLG----PGHRICTILCDSGTRYAKKQWNISFLREEGL 451
> ath:AT5G28030 cysteine synthase, putative / O-acetylserine (thiol)-lyase,
putative / O-acetylserine sulfhydrylase, putative
(EC:2.5.1.47); K01738 cysteine synthase A [EC:2.5.1.47]
Length=323
Score = 135 bits (340), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 115/334 (34%), Positives = 180/334 (53%), Gaps = 40/334 (11%)
Query 38 ILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG 97
+ + IG TP+V L+++ G ++ K E + S+KDRIA MI AE G + PG
Sbjct 11 VTELIGNTPMVYLNKI--VDGCVARIAAKLEMMEPCSSIKDRIAYSMIKDAEDKGLITPG 68
Query 98 -AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWD 156
+ LIE T GNTGIG+A + A +GY+ + +MP MS E++ I+RALG+ + T
Sbjct 69 KSTLIEATGGNTGIGLASIGASRGYKVILLMPSTMSLERRIILRALGAEVHLT------- 121
Query 157 DQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPIS 216
+SI +K ++E+ ++ K TP P Q+ NP NP
Sbjct 122 ------DISIGIKGQLEKAKEILSK--------TPGGYIP---------HQFINPENPEI 158
Query 217 HFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANP 275
H+ TGPE+ ++D++V GTGG++TG G+ +K + V V+P E ++L+
Sbjct 159 HYRTTGPEIWRDSAGKVDILVAGVGTGGTVTGTGKFLKEKNKDIKVCVVEPSESAVLSGG 218
Query 276 NEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAA 335
+PG P+L++GIG +P LD + D ++V E++ ++LL EG+LVG SSGA+
Sbjct 219 --KPG-PHLIQGIGSGEIPANLDLSIVDEIIQVTGEEAIETTKLLAIKEGLLVGISSGAS 275
Query 336 LAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTS 369
A ALK +R ++ K I ++ P G Y S
Sbjct 276 AAAALKVAKR---PENVGKLIVVIFPSGGERYLS 306
> cel:F59A7.9 hypothetical protein; K01738 cysteine synthase A
[EC:2.5.1.47]
Length=337
Score = 130 bits (328), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 98/308 (31%), Positives = 161/308 (52%), Gaps = 37/308 (12%)
Query 40 DAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG-A 98
D +G TPLV L + G+ ++ K E+L+ S+KDRIA+ M+ AE +G++ PG
Sbjct 12 DTVGNTPLVLLRNI--TKGLDARIAVKIEYLNPSCSVKDRIAKTMVDDAEKAGKIVPGKT 69
Query 99 ALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQ 158
L+E TSGN GI +A + ++GY+ + VM MS E+++++RA G+ ++ + D
Sbjct 70 VLVEGTSGNLGIALAHIGRIRGYKVILVMTTAMSIERRAMLRAYGAEVILS------DPA 123
Query 159 TSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHF 218
H + ++ E + PD A LDQ+ NP+NP +H+
Sbjct 124 EGHPGIVKKI--------------------EMLVSKLPD----AYCLDQFSNPSNPAAHY 159
Query 219 FGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNE 277
TGPE+ Q ++DMV G+GG++TG+GR ++ N V V+P E S+L+
Sbjct 160 RTTGPEIWKQTQGKVDMVCFGVGSGGTVTGVGRYLRAQKQNIGVYPVEPFESSVLSG--- 216
Query 278 QPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALA 337
P P+ + GIG +P +DR + +V +++ +R L E +L G SSGA +
Sbjct 217 FPRGPHKIHGIGAGLIPGNVDRSLFTEVFRVKSEDAMKMARRLADEEAILGGISSGANVV 276
Query 338 GALKAIER 345
A++ R
Sbjct 277 AAVELARR 284
> ath:AT3G03630 CS26; CS26; cysteine synthase; K01738 cysteine
synthase A [EC:2.5.1.47]
Length=404
Score = 121 bits (304), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 109/331 (32%), Positives = 166/331 (50%), Gaps = 52/331 (15%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAP-GAAL 100
IG TP+V L+RV G + K E + S+KDRI MI AE SG + P L
Sbjct 107 IGSTPMVYLNRV--TDGCLADIAAKLESMEPCRSVKDRIGLSMINEAENSGAITPRKTVL 164
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGS-MILRTPTKAAWDDQT 159
+EPT+GNTG+GIA VAA KGY+ + MP ++ E++ ++RALG+ ++L P K
Sbjct 165 VEPTTGNTGLGIAFVAAAKGYKLIVTMPASINIERRMLLRALGAEIVLTNPEKG------ 218
Query 160 SHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFF 219
LK +++ KE + + A + Q+ N AN HF
Sbjct 219 --------LKGAVDKA-----------------KEIVLKTKNAYMFQQFDNTANTKIHFE 253
Query 220 GTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-EGSILANPNEQ 278
TGPE+ +D+ V GTGG++TG G +K+ + VV V+P E S+++ N
Sbjct 254 TTGPEIWEDTMGNVDIFVAGIGTGGTVTGTGGFLKMMNKDIKVVGVEPSERSVISGDN-- 311
Query 279 PGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAG 338
PG ++P +LD + D KV++ E++ +R L EG+LVG SSGAA
Sbjct 312 PG-----------YLPGILDVKLLDEVFKVSNGEAIEMARRLALEEGLLVGISSGAAAVA 360
Query 339 ALKAIERMGWKDDSSKRIAMVLPDGSRNYTS 369
A+ +R +++ K I ++ P Y +
Sbjct 361 AVSLAKR---AENAGKLITVLFPSHGERYIT 388
> ath:AT3G61440 CYSC1; CYSC1 (CYSTEINE SYNTHASE C1); L-3-cyanoalanine
synthase/ cysteine synthase; K13034 L-3-cyanoalanine
synthase/ cysteine synthase [EC:2.5.1.47 4.4.1.9]
Length=272
Score = 112 bits (279), Expect = 5e-24, Method: Compositional matrix adjust.
Identities = 98/298 (32%), Positives = 145/298 (48%), Gaps = 38/298 (12%)
Query 84 MIVTAEASGRLAPG-AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRAL 142
MI AE + PG LIEPTSGN GI +A +AA+KGYR + MP S E++ MR+
Sbjct 1 MIADAEKKKLIIPGKTTLIEPTSGNMGISLAFMAAMKGYRIIMTMPSYTSLERRVTMRSF 60
Query 143 GS-MILRTPTKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQI 201
G+ ++L P K KK D D TPD
Sbjct 61 GAELVLTDPAKGMGGTV--------------------KKAYDLLDS-------TPDAFM- 92
Query 202 ACILDQYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCL 261
C Q+ NPAN HF TGPE+ +D+ V+ G+GG+++G+GR +K PN
Sbjct 93 -C--QQFANPANTQIHFDTTGPEIWEDTLGNVDIFVMGIGSGGTVSGVGRYLKSKNPNVK 149
Query 262 VVAVDPEGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLI 321
+ V+P S + N +PG P+ + G G F P +LD DV + ++V+ +++ +R L
Sbjct 150 IYGVEPAESNILN-GGKPG-PHAITGNGVGFKPEILDMDVMESVLEVSSEDAIKMARELA 207
Query 322 RLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMAE 379
EG++VG SSGA A++ + +M ++ K I + Y S + DE E
Sbjct 208 LKEGLMVGISSGANTVAAIR-LAKM--PENKGKLIVTIHASFGERYLSSVLFDELRKE 262
> ath:AT3G22460 OASA2; OASA2 (O-ACETYLSERINE (THIOL) LYASE (OAS-TL)
ISOFORM A1); catalytic/ cysteine synthase/ pyridoxal phosphate
binding; K01738 cysteine synthase A [EC:2.5.1.47]
Length=188
Score = 110 bits (276), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 77/210 (36%), Positives = 107/210 (50%), Gaps = 35/210 (16%)
Query 38 ILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPG 97
+ + IG TPLV L++V A + K E + S+KDRI MI AEA G + PG
Sbjct 12 VTELIGNTPLVYLNKV--AKDCVGHVAAKLEMMEPCSSVKDRIGYSMIADAEAKGLIKPG 69
Query 98 -AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGS-MILRTPTKAAW 155
+ LIEPTSGNTG+G+A AA KGY+ V MP MS E++ I+ A G+ +IL P K
Sbjct 70 ESVLIEPTSGNTGVGLAFTAAAKGYKLVITMPASMSIERRIILLAFGAELILTDPAKG-- 127
Query 156 DDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPI 215
+ ++ EE+ + TP+ +L Q+ NPANP
Sbjct 128 ------MKGAVAKAEEILAK-------------------TPN----GYMLQQFENPANPK 158
Query 216 SHFFGTGPELLHQCGMQLDMVVICTGTGGS 245
H+ TGPE+ G ++D V GTGG+
Sbjct 159 IHYETTGPEIWKGSGGKVDGFVSGIGTGGT 188
> sce:YGR012W Putative cysteine synthase, localized to the mitochondrial
outer membrane (EC:2.5.1.47); K01738 cysteine synthase
A [EC:2.5.1.47]
Length=393
Score = 102 bits (255), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 99/371 (26%), Positives = 171/371 (46%), Gaps = 54/371 (14%)
Query 26 FFQSHLARELP----SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIA 81
+++ R++P + + IG TPLV + + A GV+ + K E + GS KDR+A
Sbjct 33 YYKLFKTRDIPRPKEGVEELIGNTPLVKIRSLTKATGVN--IYAKLELCNPAGSAKDRVA 90
Query 82 RDMIVTAEASGRLAPG--AALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIM 139
++I TAE G L G + E TSG+TGI IA+V GYR+ +P S EK +++
Sbjct 91 LNIIKTAEELGELVRGEPGWVFEGTSGSTGISIAVVCNALGYRAHISLPDDTSLEKLALL 150
Query 140 RALGSMILRTPTKAAWDDQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDEL 199
+LG+ + + A+ D ++ + + E+++ +
Sbjct 151 ESLGATVNKVKP-ASIVDPNQYVNAAKKACNELKKSGNGIR------------------- 190
Query 200 QIACILDQYRNPANPISHFFGTGPELLHQCGMQLDMVVICTGTGGSLTGIGRRIK--LAL 257
A DQ+ N AN H+ TGPE+ HQ +D + GTGG++TG+ + +K +
Sbjct 191 --AVFADQFENEANWKVHYQTTGPEIAHQTKGNIDAFIAGCGTGGTITGVAKFLKERAKI 248
Query 258 PNCLVVAVDPEGS---------ILANPNEQPGK------PYLVEGIGYDFVPTVLD--RD 300
P C VV DP+GS ++ + E+ G +VEGIG + +
Sbjct 249 P-CHVVLADPQGSGFYNRVNYGVMYDYVEKEGTRRRHQVDTIVEGIGLNRITHNFHMGEK 307
Query 301 VADFWVKVNDAESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVL 360
D ++VND +++ ++ L +G+ VG S+ A++ + + I ++
Sbjct 308 FIDESIRVNDNQAIRMAKYLSVNDGLFVGSSTAINAVAAIQVAKTL----PHGSNIVIIA 363
Query 361 PDGSRNYTSKF 371
D + SKF
Sbjct 364 CDSGSRHLSKF 374
> cel:T25D3.3 hypothetical protein
Length=427
Score = 59.3 bits (142), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 81/362 (22%), Positives = 141/362 (38%), Gaps = 50/362 (13%)
Query 40 DAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAA 99
+ +G TP+ + G S +L+ K E S GS+K R + +++ A G + G
Sbjct 60 NLMGSTPIHVIDPPGLP---SVRLVFKNESASLTGSLKHRYSWSLMMWALLEGHVKNGTH 116
Query 100 LIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQT 159
+ E +SGNT + + + G + AV+P + K + ++R P
Sbjct 117 VFEASSGNTACSLGYMCRLLGLQFTAVVPDTIEQVKVRRIEEQEGFVVRVP--------- 167
Query 160 SHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFF 219
I+ + + E++ + + + + E P+ + H
Sbjct 168 --ISDRLIIAEQLAKSTGGFFMNQFKNAFHAEEFHESGSAS--------SKPSANLMH-- 215
Query 220 GTGPELLHQCGMQLDMVVIC-TGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILANP--- 275
EL HQ + + GTGG+L+ IGR K + +V D + SI +
Sbjct 216 ----ELFHQLNSTPPSIFVHPAGTGGTLSSIGRYAKKYGLDTEIVLADTQYSIYYDYVLN 271
Query 276 ---NEQPGKPYLV----EGIGYDFV-------PTVLDRDVADFWVKVNDAESLLCSRLLI 321
++ G + V GIGY + T LD V D +K+ D S + +
Sbjct 272 GTFSKNTGAHHWVAPGMAGIGYGAMGPAQIAETTSLDPAVIDRVLKIPDLASTAAMK-VA 330
Query 322 RLEGMLVGGSSGAALAGALKA---IERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMA 378
R G+ G S+G AL I K+ IA +L D + Y + + +W++
Sbjct 331 RDVGINGGTSTGVNFLAALHIGALIRAQNPKNQEIFTIATILADSGKYYETTYFNRKWIS 390
Query 379 EK 380
EK
Sbjct 391 EK 392
> cel:F01D4.8 hypothetical protein
Length=430
Score = 58.9 bits (141), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 82/370 (22%), Positives = 152/370 (41%), Gaps = 68/370 (18%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL-APGAAL 100
+G TPLV S G + + K E +SA ++K R A +++ A G++ + +A+
Sbjct 56 MGHTPLVKFSPPGFP---NTDIFFKNETVSATRTLKHRFAWALLLWAIVEGKVTSKTSAV 112
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTS 160
+ TSGN G A + + G AV+ ++ EK + G +++ PT
Sbjct 113 YDATSGNMGSAEAYMCTLVGIPYFAVVSEECEQEKIDNIEKFGGKVIKVPTA-------- 164
Query 161 HIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFG 220
+ + + ++N Y +K A + +++ + I++
Sbjct 165 -----------LRTTKSNEFAIENNGFYINQQKN-------AALAEEFHESGDFINY--- 203
Query 221 TGPELLHQCGMQL-----------DMVVICTGTGGSLTGIGRR-IKLALPNCLVVAVDPE 268
+ H+ +QL D V GTGG+++ +GR +K LP +V+A D +
Sbjct 204 ESSNVFHEILVQLKDNEKQTSKVPDFFVHPAGTGGTVSSVGRYVVKYGLPTKVVLA-DSQ 262
Query 269 GSILAN-------PNEQPGKPYLVEGI-----GYDFVP------TVLDRDVADFWVKVND 310
S+ + N+ K + GI GYD VP T L ++V + +K+ D
Sbjct 263 FSVYYDYVIGNKFTNQTGEKVWRAPGIAGTGYGYDIVPIQYGETTSLTKNVINEAMKMPD 322
Query 311 AESLLCSRLLIRLEGMLVGGSSGAALAGALKAIERMGWKDDSSKR---IAMVLPDGSRNY 367
++ R+L + G G S+ L + K +++ IA + D Y
Sbjct 323 IATVAAMRILNEM-GYNAGASTSLNFLVCLYKAYQTKQKSSNNEYQLTIATLAGDPGDFY 381
Query 368 TSKFVCDEWM 377
S ++ D W+
Sbjct 382 HSTYLDDGWV 391
> cel:F13B12.4 hypothetical protein
Length=435
Score = 53.5 bits (127), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 88/374 (23%), Positives = 148/374 (39%), Gaps = 68/374 (18%)
Query 42 IGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRL-APGAAL 100
+G TP+ S G + + K E +A ++K R A +++ A G++ + +A+
Sbjct 57 MGHTPMTKFSPPGFP---NADIFFKNETATATRTLKHRFAWALLLWAITEGKVTSKTSAV 113
Query 101 IEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTS 160
+ TSGNTG A + + AV+ + EK + + G I++ P
Sbjct 114 YDSTSGNTGSAEAYMCTLVNVPYYAVVADNLEKEKVKQIESFGGKIIKVP---------- 163
Query 161 HIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFG 220
+AL R K K+ D N + ++ A +++ + F+
Sbjct 164 -VAL---------RNAKAKEFADKNHGFYM------NQFGNAEKAEEFHESGD----FYF 203
Query 221 TGPELLHQCGMQL-----------DMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEG 269
+ H+ +QL D V GTGG+++ +GR + VV D +
Sbjct 204 ESTNVYHEIIVQLKKDKEQIVKIPDYFVHSAGTGGTISSVGRYVARYGAPTKVVLSDSQY 263
Query 270 SILANP------NEQPG----KPYLVEGI--GYDFVP------TVLDRDVADFWVKVNDA 311
S+ + Q G P + GI GYD P T L R+V +K+ D
Sbjct 264 SLFYDYVIGHKFTNQSGAGIWTPPGIAGIGYGYDIEPVWYGETTSLTRNVIHEAMKMPDI 323
Query 312 ESLLCSRLLIRLEGMLVGGSSG-AALAGALKAIERMGWKDDSSKRIAMVLP--DGSRNYT 368
S+ R+L +G VG S+ L KA + K R+ +V D Y
Sbjct 324 ASVATMRILDE-KGYNVGPSTSLNFLVSLYKAYQNKARKSAIKHRLTIVTLACDPGDFYR 382
Query 369 SKFVCDEWMAEKGF 382
S ++ +EW+ EK F
Sbjct 383 STYLNNEWV-EKSF 395
> hsa:63826 SRR, ILV1, ISO1; serine racemase (EC:4.3.1.17 5.1.1.18
4.3.1.18); K12235 serine racemase [EC:5.1.1.18]
Length=340
Score = 45.4 bits (106), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 82/350 (23%), Positives = 140/350 (40%), Gaps = 59/350 (16%)
Query 37 SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAP 96
+I D+I TP++ S + G + L KCE GS K R A + + +
Sbjct 18 NIRDSIHLTPVLTSSILNQLTGRN--LFFKCELFQKTGSFKIRGALNAVRSLVPDALERK 75
Query 97 GAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWD 156
A++ +SGN G + A ++G + V+P+ K+ ++A G+ I+ D
Sbjct 76 PKAVVTHSSGNHGQALTYAAKLEGIPAYIVVPQTAPDCKKLAIQAYGASIVYCEPS---D 132
Query 157 DQTSHIALSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPIS 216
+ ++A R+ EE E P +E A I Q
Sbjct 133 ESRENVAK--RVTEETE------------GIMVHPNQEP------AVIAGQ--------- 163
Query 217 HFFGT-GPELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDP-------- 267
GT E+L+Q + +D +V+ G GG L GI +K P+ V A +P
Sbjct 164 ---GTIALEVLNQVPL-VDALVVPVGGGGMLAGIAITVKALKPSVKVYAAEPSNADDCYQ 219
Query 268 ---EGSILANPNEQPGKPY---LVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSRLLI 321
+G ++ PN P + + IG + P + RD+ D V + E ++L+
Sbjct 220 SKLKGKLM--PNLYPPETIADGVKSSIGLNTWPII--RDLVDDIFTVTEDEIKCATQLVW 275
Query 322 RLEGMLVGGSSGAALAGALKAIERMGWKDDSSKRIAMVLPDGSRNYTSKF 371
+L+ ++G +A L + K I +VL G+ + TS
Sbjct 276 ERMKLLIEPTAGVGVAAVLS--QHFQTVSPEVKNICIVLSGGNVDLTSSI 323
> hsa:781 CACNA2D1, CACNA2, CACNL2A, CCHL2A; calcium channel,
voltage-dependent, alpha 2/delta subunit 1; K04858 voltage-dependent
calcium channel alpha-2/delta-1
Length=1091
Score = 41.2 bits (95), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 4/86 (4%)
Query 219 FGTGPELLHQCGMQLDMVVICTGTGGSLTGIGR---RIKLALPNCLVVAVDPEGSILANP 275
F + ++ ++ +++ G SL I R R L PN A+DP G +L +P
Sbjct 467 FNITGQFENKTNLKNQLILGVMGVDVSLEDIKRLTPRFTLC-PNGYYFAIDPNGYVLLHP 525
Query 276 NEQPGKPYLVEGIGYDFVPTVLDRDV 301
N QP P E + DF+ L+ D+
Sbjct 526 NLQPKNPKSQEPVTLDFLDAELENDI 551
> ath:AT4G11640 ATSR; ATSR (ARABIDOPSIS THALIANA SERINE RACEMASE);
serine racemase (EC:4.3.1.18); K12235 serine racemase [EC:5.1.1.18]
Length=331
Score = 41.2 bits (95), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 55/237 (23%), Positives = 102/237 (43%), Gaps = 40/237 (16%)
Query 45 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALIEPT 104
TP++ + + +G S L KCE L GG+ K R A + +++ +A + A G ++ +
Sbjct 29 TPVLTSESLNSISGRS--LFFKCECLQKGGAFKFRGACNAVLSLDAE-QAAKG--VVTHS 83
Query 105 SGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWDDQTSHIAL 164
SGN ++L A ++G + V+PK A K + ++R K W + T
Sbjct 84 SGNHAAALSLAAKIQGIPAYIVVPK--GAPKCKV-----DNVIRYGGKVIWSEAT----- 131
Query 165 SIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFGTGPE 224
+ +EE+ + + + +L N IS E
Sbjct 132 -MSSREEIASKVLQ---------------------ETGSVLIHPYNDGRIISGQGTIALE 169
Query 225 LLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILANPNEQPGK 281
LL Q ++D +V+ GG ++G+ K P+ ++A +P+G+ A ++ GK
Sbjct 170 LLEQI-QEIDAIVVPISGGGLISGVALAAKSIKPSIRIIAAEPKGADDAAQSKVAGK 225
> dre:100331432 serine racemase-like
Length=247
Score = 40.0 bits (92), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 44/112 (39%), Gaps = 6/112 (5%)
Query 45 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALIEPT 104
TPL+ + SC + K E + GS K R A GR G + +
Sbjct 6 TPLIPWCQTTLPPHTSCNIHIKMENMQRTGSFKIR------GVANQFGRRLKGDHFVTMS 59
Query 105 SGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPTKAAWD 156
SGN G A G++ VM + + ++++LG + R PT D
Sbjct 60 SGNYGKAFAFACKHYGFKGKVVMLENAPISRSELIKSLGVEVERVPTTQLMD 111
> dre:561434 cacna2d1a, si:ch211-251k4.1; calcium channel, voltage-dependent,
alpha 2/delta subunit 1a; K04858 voltage-dependent
calcium channel alpha-2/delta-1
Length=1069
Score = 38.9 bits (89), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 39/141 (27%), Positives = 62/141 (43%), Gaps = 14/141 (9%)
Query 258 PNCLVVAVDPEGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLL-C 316
PN A+DP G +L +PN QP E + DF LD ++ D VKV+ S++
Sbjct 508 PNGYYFAIDPNGYVLLHPNLQPKDLKSQEPVTLDF----LDAEMEDE-VKVDIRRSMIDG 562
Query 317 SRLLIRLEGMLVGGSSGAALAGALKAIERMGWK--DDSSKRIAMVLPDGSRNYTSKFVCD 374
R IR ++ G W D+ +A+VLPD S +Y + D
Sbjct 563 ERSQIRFHTLMKSQDERYIDKG----FRTYTWSPVTDADYSLALVLPDYSMSYIKATIGD 618
Query 375 EWMAEKGFLERSELSRQYEQY 395
+ + F E ++ + +E+Y
Sbjct 619 T-ITQAKFSESLQVDK-FEEY 637
> mmu:27364 Srr, Srs; serine racemase (EC:4.3.1.17 5.1.1.18 4.3.1.18);
K12235 serine racemase [EC:5.1.1.18]
Length=339
Score = 37.7 bits (86), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 52/111 (46%), Gaps = 2/111 (1%)
Query 37 SILDAIGCTPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAP 96
+I D+I TP++ S + AG + L KCE GS K R A + I
Sbjct 18 NIQDSIHLTPVLTSSILNQIAGRN--LFFKCELFQKTGSFKIRGALNAIRGLIPDTPEEK 75
Query 97 GAALIEPTSGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMIL 147
A++ +SGN G + A ++G + V+P+ K+ ++A G+ I+
Sbjct 76 PKAVVTHSSGNHGQALTYAAKLEGIPAYIVVPQTAPNCKKLAIQAYGASIV 126
> cel:T01H8.2 hypothetical protein
Length=317
Score = 37.7 bits (86), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 55/234 (23%), Positives = 83/234 (35%), Gaps = 46/234 (19%)
Query 45 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALIEPT 104
T ++ + G +L KCE L GS K R A + + A+ +I +
Sbjct 24 TLVITSENIDEKVGNGTHVLFKCEHLQKTGSFKARGALNSAILAKEKN----AKGMIAHS 79
Query 105 SGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRAL-GSMILRTPTKAAWDDQTSHIA 163
SGN G +A A G V+PK K MR +++ PT TS +
Sbjct 80 SGNHGQALAWAAQKIGLPCTIVVPKNAPISKIEGMREYNANIVFCEPT------VTSRES 133
Query 164 LSIRLKEEMERQRKEKKKDDSNDCYETPEKETPDELQIACILDQYRNPANPISHFFGTGP 223
+ L E++E C E P N +S G
Sbjct 134 VCADLTEKLEYY-----------CVE---------------------PYNCVSMINGHSS 161
Query 224 ---ELLHQCGMQLDMVVICTGTGGSLTGIGRRIKLALPNCLVVAVDPEGSILAN 274
E+L Q G ++D + + G GG + + I P+ V V P L+N
Sbjct 162 VAFEILEQVGNEIDSIFLSVGGGGLASSVAFLIGNLRPDIEVYLVQPAQKELSN 215
> xla:397727 sox13, Sry, sox12, xSox12, xsox13; SRY (sex determining
region Y)-box 13; K09269 transcription factor SOX5/6/13
(SOX group D)
Length=567
Score = 35.0 bits (79), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 38/179 (21%), Positives = 71/179 (39%), Gaps = 27/179 (15%)
Query 232 QLDMVVICTGTGGSLTGIGRRIKLALP-------NCLVVAVDPEGSILANPNEQPGKPYL 284
+ M + + GG++ K LP + + A+ +L N P +
Sbjct 290 EFRMSPVGSQCGGTMDSASSSQKANLPLGFLGEGDAITKAIQEACQLLHGQNTSPE--HC 347
Query 285 VEGIGYDFVPTVLDRDVADFWVKVNDAESLL-CSRLLIRLEGMLVGGSSGAALAGALKAI 343
+ + + T+ ++++ D + SLL CS M + GS G + + A
Sbjct 348 QQNYRKELLDTLPEKNIQDVTSLHHTEASLLRCS--------MDIDGSRGNHIKRPMNAF 399
Query 344 ERMGWKDDSSKRIAMVLPDGSRNYTSKFVCDEWMA------EKGFLERSELSRQY-EQY 395
M W D ++I PD + SK + W + + + E++ LSRQ+ E+Y
Sbjct 400 --MVWAKDERRKILQAFPDMHNSSISKILGSRWKSMSNGEKQPYYEEQARLSRQHLERY 456
> dre:100333639 calcium channel, voltage-dependent, alpha 2/delta
subunit 2-like; K04859 voltage-dependent calcium channel
alpha-2/delta-2
Length=1052
Score = 35.0 bits (79), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 54/122 (44%), Gaps = 30/122 (24%)
Query 259 NCLVVAVDPEGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSR 318
N + A+DP G +L +PN QP + L E + DF LD +V D S+
Sbjct 455 NGYIFAIDPNGYVLMHPNLQPREEDLPEQVTLDF----LDAEVED------------SSK 498
Query 319 LLIRLEGMLVGGSSGAALAGALKAIE----------RM-GWK--DDSSKRIAMVLPDGSR 365
IR + M+ G S + +K+++ RM W +D+ + +VLP S
Sbjct 499 QEIRRQ-MIDGRSGDMTIKTLIKSMDEASRYIDDVTRMYTWTPINDTDYSLGLVLPPYSE 557
Query 366 NY 367
+Y
Sbjct 558 HY 559
> dre:568759 novel protein similar to vertebrate calcium channel
voltage-dependent alpha 2 delta subunit 2 (CACNA2D2); K04859
voltage-dependent calcium channel alpha-2/delta-2
Length=1052
Score = 35.0 bits (79), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 54/122 (44%), Gaps = 30/122 (24%)
Query 259 NCLVVAVDPEGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVNDAESLLCSR 318
N + A+DP G +L +PN QP + L E + DF LD +V D S+
Sbjct 455 NGYIFAIDPNGYVLMHPNLQPREEDLPEQVTLDF----LDAEVED------------SSK 498
Query 319 LLIRLEGMLVGGSSGAALAGALKAIE----------RM-GWK--DDSSKRIAMVLPDGSR 365
IR + M+ G S + +K+++ RM W +D+ + +VLP S
Sbjct 499 QEIRRQ-MIDGRSGDMTIKTLIKSMDEASRYIDDVTRMYTWTPINDTDYSLGLVLPPYSE 557
Query 366 NY 367
+Y
Sbjct 558 HY 559
> mmu:232078 Thnsl2, BC051244, MGC59076, TSH2; threonine synthase-like
2 (bacterial) (EC:4.2.3.-)
Length=483
Score = 33.1 bits (74), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 12/57 (21%)
Query 217 HFFGTGPELLHQCGMQLDM-------VVICTGTGGSLTGIGRRIKLALPNCLVVAVD 266
HFF QC LD VV+ TG GG+L K+ LP CLVVAV+
Sbjct 235 HFFA-----YFQCTPSLDTHPLPTVEVVVPTGAGGNLAAGCIAQKMGLPICLVVAVN 286
> dre:100331498 serine racemase-like
Length=323
Score = 32.3 bits (72), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 25/107 (23%), Positives = 42/107 (39%), Gaps = 6/107 (5%)
Query 45 TPLVCLSRVGTAAGVSCQLLGKCEFLSAGGSMKDRIARDMIVTAEASGRLAPGAALIEPT 104
TP++ S+ SC + K E + GS K R A + G + +
Sbjct 31 TPMIPWSQTTLPLHTSCNIHIKLENMQRTGSFKIR------GVANQFAKRHKGDHFVTMS 84
Query 105 SGNTGIGIALVAAVKGYRSVAVMPKKMSAEKQSIMRALGSMILRTPT 151
+GN G + G + VMP+ + +++ LG + R PT
Sbjct 85 AGNYGKAFSYACKHYGSKGKVVMPETAPISRSVLIQNLGVEVERVPT 131
> cel:F59A7.7 hypothetical protein
Length=162
Score = 32.3 bits (72), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 46/92 (50%), Gaps = 4/92 (4%)
Query 251 RRIKLALPNCLVVAVDP-EGSILANPNEQPGKPYLVEGIGYDFVPTVLDRDVADFWVKVN 309
R ++ N V V+P E S+L+ P P+ + GIG +P +DR + ++V
Sbjct 17 RYLRAQKQNIGVYPVEPFESSVLSG---FPRGPHKIHGIGAGLIPGNVDRSLFTEVLRVK 73
Query 310 DAESLLCSRLLIRLEGMLVGGSSGAALAGALK 341
+++ +R L E +L SSGA + A++
Sbjct 74 SEDAMKMARRLADEEAILGEISSGANVVAAVE 105
Lambda K H
0.316 0.132 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 27194813240
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40