bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1907_orf2
Length=190
Score E
Sequences producing significant alignments: (Bits) Value
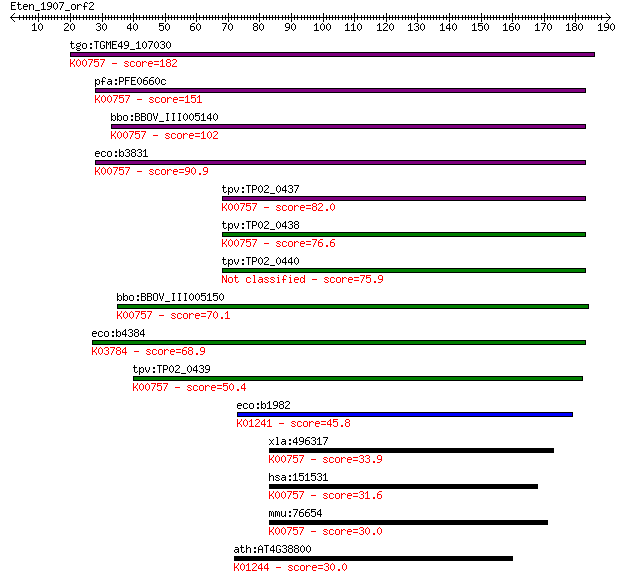
tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3)... 182 5e-46
pfa:PFE0660c purine nucleotide phosphorylase, putative (EC:2.4... 151 1e-36
bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC... 102 7e-22
eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.... 90.9 2e-18
tpv:TP02_0437 purine nucleoside phosphorylase; K00757 uridine ... 82.0 1e-15
tpv:TP02_0438 purine nucleoside phosphorylase; K00757 uridine ... 76.6 5e-14
tpv:TP02_0440 purine nucleoside phosphorylase 75.9 9e-14
bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uri... 70.1 4e-12
eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosph... 68.9 9e-12
tpv:TP02_0439 hypothetical protein; K00757 uridine phosphoryla... 50.4 3e-06
eco:b1982 amn, ECK1977, JW1963; AMP nucleosidase (EC:3.2.2.4);... 45.8 1e-04
xla:496317 upp2; uridine phosphorylase 2 (EC:2.4.2.3); K00757 ... 33.9 0.38
hsa:151531 UPP2, UDRPASE2, UP2, UPASE2; uridine phosphorylase ... 31.6 1.8
mmu:76654 Upp2, 1700124F02Rik, AI266885, UDRPASE2, UP2, UPASE2... 30.0 5.6
ath:AT4G38800 ATMTN1; ATMTN1; catalytic/ methylthioadenosine n... 30.0 5.6
> tgo:TGME49_107030 purine nucleoside phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=247
Score = 182 bits (463), Expect = 5e-46, Method: Compositional matrix adjust.
Identities = 85/166 (51%), Positives = 116/166 (69%), Gaps = 0/166 (0%)
Query 20 MEGVEVQPHIQLRRGQVQPVVLTMGDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQK 79
M+G+EVQPHI+LR+ V+PVV+ +GDP R +EVA +C+ + L +NRE++S VV D Q
Sbjct 1 MQGMEVQPHIRLRKEDVEPVVIIVGDPARTEEVANMCEKKQELAYNREYRSFRVVYDSQP 60
Query 80 FTVISHGIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVC 139
TVISHGIGC + I EEL LGAK IIRAGTCGS+KP +K+GD+CV +AA + +
Sbjct 61 ITVISHGIGCPGTSIAIEELAYLGAKVIIRAGTCGSLKPKTLKQGDVCVTYAAVNETGLI 120
Query 140 DLYIPPRMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFYGHG 185
+P P VATP V+Q+L+++ K+LGI GI +T FY +G
Sbjct 121 SNILPEGFPCVATPHVYQALMDAAKELGIEAASGIGVTQDYFYQNG 166
> pfa:PFE0660c purine nucleotide phosphorylase, putative (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=245
Score = 151 bits (381), Expect = 1e-36, Method: Compositional matrix adjust.
Identities = 69/155 (44%), Positives = 104/155 (67%), Gaps = 0/155 (0%)
Query 28 HIQLRRGQVQPVVLTMGDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQKFTVISHGI 87
H+++ + Q+ PVVL +GDP RVD++ +CDS L +NRE+KS GQKF +SHG+
Sbjct 7 HLKISKEQITPVVLVVGDPGRVDKIKVVCDSYVDLAYNREYKSVECHYKGQKFLCVSHGV 66
Query 88 GCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVCDLYIPPRM 147
G +CFEEL + GAK IIRAG+CGS++PD +KRGDIC+ AA R++ V L I
Sbjct 67 GSAGCAVCFEELCQNGAKVIIRAGSCGSLQPDLIKRGDICICNAAVREDRVSHLLIHGDF 126
Query 148 PAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
PAV V+ +L + ++L +P+ GI++++ ++Y
Sbjct 127 PAVGDFDVYDTLNKCAQELNVPVFNGISVSSDMYY 161
> bbo:BBOV_III005140 17.m07459; phosphorylase family protein (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 102 bits (254), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 53/150 (35%), Positives = 81/150 (54%), Gaps = 0/150 (0%)
Query 33 RGQVQPVVLTMGDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQKFTVISHGIGCCSS 92
+G+ V + GD KR + +K A + DG+ + SHG+G +
Sbjct 16 KGRDYKVAIICGDHKRFEIFKEHSTGYRDFGTYLCWKGAELFYDGEYILLASHGVGSHPT 75
Query 93 MICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVCDLYIPPRMPAVAT 152
+ ELI+LG KCIIR+GTCG+ +PD GDIC+ + A R++ C PAVA
Sbjct 76 YVVVRELIELGVKCIIRSGTCGTYRPDIRSTGDICICYGAIRNDSSCLGDADLAYPAVAH 135
Query 153 PRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
P VF++L ++ + LGIP G++ ++ LFY
Sbjct 136 PNVFEALRDASEALGIPTHLGMSYSSDLFY 165
> eco:b3831 udp, ECK3825, JW3808; uridine phosphorylase (EC:2.4.2.3);
K00757 uridine phosphorylase [EC:2.4.2.3]
Length=253
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 53/157 (33%), Positives = 82/157 (52%), Gaps = 3/157 (1%)
Query 28 HIQLRRGQVQPVVLTM--GDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQKFTVISH 85
H+ L + +Q L + GDP RV+++AAL D L +REF + DG+ V S
Sbjct 8 HLGLTKNDLQGATLAIVPGDPDRVEKIAALMDKPVKLASHREFTTWRAELDGKPVIVCST 67
Query 86 GIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVCDLYIPP 145
GIG S+ I EEL +LG + +R GT G+++P + GD+ V A+ R + + P
Sbjct 68 GIGGPSTSIAVEELAQLGIRTFLRIGTTGAIQP-HINVGDVLVTTASVRLDGASLHFAPL 126
Query 146 RMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
PAVA +L+E+ K +G G+ ++ FY
Sbjct 127 EFPAVADFECTTALVEAAKSIGATTHVGVTASSDTFY 163
> tpv:TP02_0437 purine nucleoside phosphorylase; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=256
Score = 82.0 bits (201), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 66/115 (57%), Gaps = 0/115 (0%)
Query 68 FKSANVVKDGQKFTVISHGIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDIC 127
+K A V DG+ ++ HG+ ++ +EL+++G K +IR G CG+ KP VK GD+
Sbjct 51 YKIAEVEVDGEMILLVYHGVSGNAASPIMKELLEMGVKIVIRCGKCGTYKPSTVKSGDLS 110
Query 128 VPFAAARDNDVCDLYIPPRMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
+ +A+ RD+ I + PAVA V L+++ ++LGI GI ++ LFY
Sbjct 111 ITYASVRDDTSSMAEIDVKYPAVADFDVIDCLVDTAEELGIDASLGINYSSDLFY 165
> tpv:TP02_0438 purine nucleoside phosphorylase; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=254
Score = 76.6 bits (187), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 36/115 (31%), Positives = 61/115 (53%), Gaps = 0/115 (0%)
Query 68 FKSANVVKDGQKFTVISHGIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDIC 127
+K A V DG+ ++ HG+ ++ +EL+++G K +IR G CG+ P VK GD+
Sbjct 51 YKIAEVEVDGEMILIVYHGVSGTAASPIMKELLEMGVKIVIRCGICGANNPSTVKSGDLS 110
Query 128 VPFAAARDNDVCDLYIPPRMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
+ +A+ RD+ I + PAVA V L+++ + G+ GI T +Y
Sbjct 111 ITYASVRDDSSSLAEIDVKYPAVADYDVIDCLVDTSDEFGVKASLGINYTTDSYY 165
> tpv:TP02_0440 purine nucleoside phosphorylase
Length=254
Score = 75.9 bits (185), Expect = 9e-14, Method: Compositional matrix adjust.
Identities = 40/115 (34%), Positives = 61/115 (53%), Gaps = 0/115 (0%)
Query 68 FKSANVVKDGQKFTVISHGIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDIC 127
+K A + DG ++ HG+ + EL+++G K +IR G GS KP QVK GDI
Sbjct 51 YKIAELEVDGVMILLVYHGVSGNAVSPILTELLEMGVKIVIRCGKAGSFKPSQVKSGDIY 110
Query 128 VPFAAARDNDVCDLYIPPRMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
+ +A+ RD+ I + PAVA V L+++ ++LGI GI + FY
Sbjct 111 ICYASMRDDSSSLAEIDVKYPAVADYDVIDCLVDTAEELGIKTNLGINYSTDSFY 165
> bbo:BBOV_III005150 17.m07460; hypothetical protein; K00757 uridine
phosphorylase [EC:2.4.2.3]
Length=260
Score = 70.1 bits (170), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 69/149 (46%), Gaps = 0/149 (0%)
Query 35 QVQPVVLTMGDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQKFTVISHGIGCCSSMI 94
++ PV + +GDPK ++ +A+L E + S + +GQ F +S G G +
Sbjct 21 RIHPVAIVVGDPKWLEMLASLATRQEFFARYKSLSSMELEFNGQTFFALSFGFGATNLHR 80
Query 95 CFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVCDLYIPPRMPAVATPR 154
EL G KC+I S+ P +V +C+ +AA R + + + PA+A P
Sbjct 81 LLTELAVFGVKCVILITNTFSLLPGRVPANALCITYAACRGDYASTIEVDLAYPAIAHPD 140
Query 155 VFQSLLESGKQLGIPLRQGIALTNGLFYG 183
++L ++ ++L + LTN Y
Sbjct 141 ATRALRKAAEELKLDYHLTRTLTNDYAYN 169
> eco:b4384 deoD, ECK4376, JW4347, pup; purine-nucleoside phosphorylase
(EC:2.4.2.1); K03784 purine-nucleoside phosphorylase
[EC:2.4.2.1]
Length=239
Score = 68.9 bits (167), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 51/159 (32%), Positives = 75/159 (47%), Gaps = 4/159 (2%)
Query 27 PHIQLRRGQVQPVVLTMGDPKRVDEVA-ALCDSAEPLTFNREFKSANVVKDGQKFTVISH 85
PHI G VVL GDP R +A + A + R G+K +V+ H
Sbjct 4 PHINAEMGDFADVVLMPGDPLRAKYIAETFLEDAREVNNVRGMLGFTGTYKGRKISVMGH 63
Query 86 GIGCCSSMICFEELIK-LGAKCIIRAGTCGSMKPDQVKRGDICVPFAAARDNDVCDL-YI 143
G+G S I +ELI G K IIR G+CG++ P VK D+ + A D+ V + +
Sbjct 64 GMGIPSCSIYTKELITDFGVKKIIRVGSCGAVLP-HVKLRDVVIGMGACTDSKVNRIRFK 122
Query 144 PPRMPAVATPRVFQSLLESGKQLGIPLRQGIALTNGLFY 182
A+A + ++ +++ K LGI R G + LFY
Sbjct 123 DHDFAAIADFDMVRNAVDAAKALGIDARVGNLFSADLFY 161
> tpv:TP02_0439 hypothetical protein; K00757 uridine phosphorylase
[EC:2.4.2.3]
Length=271
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 38/144 (26%), Positives = 61/144 (42%), Gaps = 2/144 (1%)
Query 40 VLTMGDPKRVDEVAALCDSAEPLTFNREFKSANVVKDGQKFTVISHGIGCCSSMICFEEL 99
V+ GD D + DS + KS + G+K + S G G E
Sbjct 41 VIVFGDHSWFDVFLPMTDSYIEFGHYKNMKSVEISYKGRKILLYSFGFGFVDLNRVVTES 100
Query 100 IKLGAKCIIRAGTCGSMKPD-QVKRGDICVPFAAARDNDVCDLYIPPRMPAVATPRVFQS 158
LGA +I + P Q+K DI +A+ R+ + + + R PAVA P + ++
Sbjct 101 WFLGASNMIFLNFSIPLLPSSQIKDKDIFTIYASCRNENGTFIEVDYRYPAVAHPTIVRA 160
Query 159 LLESGKQLGIPLR-QGIALTNGLF 181
L S + LG L+ Q + +G +
Sbjct 161 LRTSSETLGYTLKPQKSVVVDGYY 184
> eco:b1982 amn, ECK1977, JW1963; AMP nucleosidase (EC:3.2.2.4);
K01241 AMP nucleosidase [EC:3.2.2.4]
Length=484
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 53/114 (46%), Gaps = 9/114 (7%)
Query 73 VVKDGQKFTVISHGIGCCSSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDICVPFAA 132
+ DGQ T+++ G+G ++ + L L + G CG ++ Q GD + A
Sbjct 266 ITADGQGITLVNIGVGPSNAKTICDHLAVLRPDVWLMIGHCGGLRESQAI-GDYVLAHAY 324
Query 133 ARDNDVCDLYIPPRMPAVATPRVFQSLLES--------GKQLGIPLRQGIALTN 178
RD+ V D +PP +P + V ++L ++ G+++ LR G +T
Sbjct 325 LRDDHVLDAVLPPDIPIPSIAEVQRALYDATKLVSGRPGEEVKQRLRTGTVVTT 378
> xla:496317 upp2; uridine phosphorylase 2 (EC:2.4.2.3); K00757
uridine phosphorylase [EC:2.4.2.3]
Length=316
Score = 33.9 bits (76), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 34/98 (34%), Positives = 42/98 (42%), Gaps = 19/98 (19%)
Query 83 ISHGIGCCSSMICFEELIKL--GAKC----IIRAGTCG--SMKPDQVKRGDICVPFAAAR 134
ISHG+G S I ELIKL AKC IIR GT G ++P V D+ V
Sbjct 112 ISHGMGIPSISIMLHELIKLLHHAKCRDVTIIRIGTSGGIGIEPGTVVITDVAV------ 165
Query 135 DNDVCDLYIPPRMPAVATPRVFQSLLESGKQLGIPLRQ 172
D + P+ V +V E K+L L Q
Sbjct 166 -----DSFFRPQFEQVVLDKVVVRKTELHKELAEDLLQ 198
> hsa:151531 UPP2, UDRPASE2, UP2, UPASE2; uridine phosphorylase
2 (EC:2.4.2.3); K00757 uridine phosphorylase [EC:2.4.2.3]
Length=374
Score = 31.6 bits (70), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 32/93 (34%), Positives = 38/93 (40%), Gaps = 19/93 (20%)
Query 83 ISHGIGCCSSMICFEELIKL--GAKC----IIRAGTCG--SMKPDQVKRGDICVPFAAAR 134
ISHG+G S I ELIKL A+C IIR GT G + P V DI V
Sbjct 169 ISHGMGIPSISIMLHELIKLLHHARCCDVTIIRIGTSGGIGIAPGTVVITDIAV------ 222
Query 135 DNDVCDLYIPPRMPAVATPRVFQSLLESGKQLG 167
D + PR V + E K+L
Sbjct 223 -----DSFFKPRFEQVILDNIVTRSTELDKELS 250
> mmu:76654 Upp2, 1700124F02Rik, AI266885, UDRPASE2, UP2, UPASE2;
uridine phosphorylase 2 (EC:2.4.2.3); K00757 uridine phosphorylase
[EC:2.4.2.3]
Length=320
Score = 30.0 bits (66), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 36/94 (38%), Gaps = 15/94 (15%)
Query 83 ISHGIGCCSSMICFEELIKL--GAKC----IIRAGTCGSMKPDQVKRGDICVPFAAARDN 136
+SHG+G S I ELIKL A C IIR GT G + P + +
Sbjct 112 VSHGMGIPSISIMLHELIKLLHHAHCCDVTIIRIGTSGGIG---------IAPGSVVITD 162
Query 137 DVCDLYIPPRMPAVATPRVFQSLLESGKQLGIPL 170
D + PR V V E K+L L
Sbjct 163 TAVDSFFKPRFEQVILDNVVTRSTELDKELANDL 196
> ath:AT4G38800 ATMTN1; ATMTN1; catalytic/ methylthioadenosine
nucleosidase (EC:3.2.2.16); K01244 5'-methylthioadenosine nucleosidase
[EC:3.2.2.16]
Length=267
Score = 30.0 bits (66), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 39/94 (41%), Gaps = 6/94 (6%)
Query 72 NVVKDGQKFTVISHGIGCC-SSMICFEELIKLGAKCIIRAGTCGSMKPDQVKRGDI-CVP 129
NVV G+ + +G +S+I F + L II AGTCG K GD+ V
Sbjct 75 NVVCPGRDAALGIDSVGTVPASLITFASIQALKPDIIINAGTCGGFKVKGANIGDVFLVS 134
Query 130 FAAARDNDV----CDLYIPPRMPAVATPRVFQSL 159
D + DLY A +TP + + L
Sbjct 135 DVVFHDRRIPIPMFDLYGVGLRQAFSTPNLLKEL 168
Lambda K H
0.324 0.141 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5429324208
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40