bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1873_orf1
Length=172
Score E
Sequences producing significant alignments: (Bits) Value
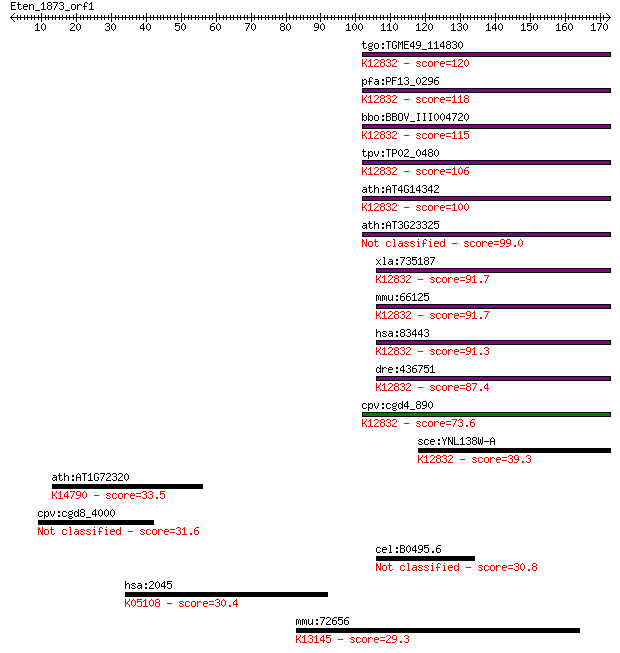
tgo:TGME49_114830 splicing factor 3b subunit 10, putative ; K1... 120 2e-27
pfa:PF13_0296 splicing factor 3b subunit, putative; K12832 spl... 118 9e-27
bbo:BBOV_III004720 17.m07423; splicing factor 3B subunit 10 (S... 115 7e-26
tpv:TP02_0480 hypothetical protein; K12832 splicing factor 3B ... 106 4e-23
ath:AT4G14342 pre-mRNA splicing factor 10 kDa subunit, putativ... 100 3e-21
ath:AT3G23325 splicing factor, putative 99.0 8e-21
xla:735187 sf3b5, MGC131202; splicing factor 3b, subunit 5, 10... 91.7 1e-18
mmu:66125 Sf3b5, 10kDa, 1110005L13Rik, AU043053, Sf3b10; splic... 91.7 1e-18
hsa:83443 SF3B5, MGC3133, SF3b10, Ysf3; splicing factor 3b, su... 91.3 1e-18
dre:436751 sf3b5, si:ch211-8a9.5, zgc:92874; splicing factor 3... 87.4 2e-17
cpv:cgd4_890 hypothetical protein ; K12832 splicing factor 3B ... 73.6 3e-13
sce:YNL138W-A YSF3, RCP10; Component of the SF3b subcomplex of... 39.3 0.007
ath:AT1G72320 APUM23; APUM23 (Arabidopsis Pumilio 23); RNA bin... 33.5 0.34
cpv:cgd8_4000 hypothetical protein 31.6 1.4
cel:B0495.6 hypothetical protein 30.8 2.3
hsa:2045 EPHA7, EHK3, HEK11; EPH receptor A7 (EC:2.7.10.1); K0... 30.4 2.9
mmu:72656 Ints8, 2810013E07Rik, AV063769, D130008D20Rik; integ... 29.3 6.5
> tgo:TGME49_114830 splicing factor 3b subunit 10, putative ;
K12832 splicing factor 3B subunit 5
Length=108
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 52/71 (73%), Positives = 62/71 (87%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
M+ DRF+I +QLQHLQSKY GTGNA TT+L+W +IQRDTLASHVGHYSRLAYFAV EN
Sbjct 1 MAGYDRFSIQAQLQHLQSKYQGTGNAQTTKLEWATSIQRDTLASHVGHYSRLAYFAVVEN 60
Query 162 ESIRRIRYRCL 172
E+++R+RYR L
Sbjct 61 ENVKRLRYRFL 71
> pfa:PF13_0296 splicing factor 3b subunit, putative; K12832 splicing
factor 3B subunit 5
Length=86
Score = 118 bits (296), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 53/71 (74%), Positives = 60/71 (84%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
MS DRF IH+QL+HLQSKY G+G+ADT+R +W NI RDTLASHVGHYSRLAYFAV EN
Sbjct 1 MSTFDRFNIHAQLEHLQSKYQGSGHADTSRWEWLTNIHRDTLASHVGHYSRLAYFAVVEN 60
Query 162 ESIRRIRYRCL 172
E I +IRYRCL
Sbjct 61 EPIAKIRYRCL 71
> bbo:BBOV_III004720 17.m07423; splicing factor 3B subunit 10
(SF3b10); K12832 splicing factor 3B subunit 5
Length=87
Score = 115 bits (288), Expect = 7e-26, Method: Compositional matrix adjust.
Identities = 50/71 (70%), Positives = 60/71 (84%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
MS DRF IH+QL+HLQSKY GTG+ + T+ +W +NIQRDTLASH GHY+RLAYFA+ EN
Sbjct 1 MSGYDRFNIHAQLEHLQSKYQGTGHVNNTKWEWALNIQRDTLASHAGHYTRLAYFAICEN 60
Query 162 ESIRRIRYRCL 172
ESI RIR+RCL
Sbjct 61 ESISRIRHRCL 71
> tpv:TP02_0480 hypothetical protein; K12832 splicing factor 3B
subunit 5
Length=94
Score = 106 bits (265), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 45/71 (63%), Positives = 59/71 (83%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
MS+ DRF IH+QL+HLQSKY GTG+ D T+ +W +NIQRDTL+SH GHY+RLAYF++ EN
Sbjct 1 MSSYDRFNIHAQLEHLQSKYQGTGHVDNTKWEWVLNIQRDTLSSHCGHYTRLAYFSIVEN 60
Query 162 ESIRRIRYRCL 172
E + RI++R L
Sbjct 61 EPVFRIKHRFL 71
> ath:AT4G14342 pre-mRNA splicing factor 10 kDa subunit, putative;
K12832 splicing factor 3B subunit 5
Length=87
Score = 100 bits (248), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 43/71 (60%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
M DRF I+SQL+HLQ+KY GTG+AD +R +W +NIQRD+ AS++GHY L+YFA+AEN
Sbjct 1 MQASDRFNINSQLEHLQAKYVGTGHADLSRFEWAVNIQRDSYASYIGHYPMLSYFAIAEN 60
Query 162 ESIRRIRYRCL 172
ESI R RY +
Sbjct 61 ESIGRERYNFM 71
> ath:AT3G23325 splicing factor, putative
Length=87
Score = 99.0 bits (245), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 43/71 (60%), Positives = 56/71 (78%), Gaps = 0/71 (0%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAEN 161
M DRF I+SQL+HLQ+KY GTG+AD +R +W +NIQRD+ AS++GHY L+YFA+AEN
Sbjct 1 MQASDRFNINSQLEHLQAKYVGTGHADLSRFEWTVNIQRDSYASYIGHYPMLSYFAIAEN 60
Query 162 ESIRRIRYRCL 172
ESI R RY +
Sbjct 61 ESIGRERYNFM 71
> xla:735187 sf3b5, MGC131202; splicing factor 3b, subunit 5,
10kDa; K12832 splicing factor 3B subunit 5
Length=86
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 40/67 (59%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+TIHSQL+HLQSKY GTG+ADTT+ +W +N RD+ S++GH+ L YFAVAENES
Sbjct 3 DRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAVAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> mmu:66125 Sf3b5, 10kDa, 1110005L13Rik, AU043053, Sf3b10; splicing
factor 3b, subunit 5; K12832 splicing factor 3B subunit
5
Length=86
Score = 91.7 bits (226), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/67 (58%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+TIHSQL+HLQSKY GTG+ADTT+ +W +N RD+ S++GH+ L YFA+AENES
Sbjct 3 DRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAIAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> hsa:83443 SF3B5, MGC3133, SF3b10, Ysf3; splicing factor 3b,
subunit 5, 10kDa; K12832 splicing factor 3B subunit 5
Length=86
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 39/67 (58%), Positives = 53/67 (79%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+TIHSQL+HLQSKY GTG+ADTT+ +W +N RD+ S++GH+ L YFA+AENES
Sbjct 3 DRYTIHSQLEHLQSKYIGTGHADTTKWEWLVNQHRDSYCSYMGHFDLLNYFAIAENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> dre:436751 sf3b5, si:ch211-8a9.5, zgc:92874; splicing factor
3b, subunit 5; K12832 splicing factor 3B subunit 5
Length=86
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 36/67 (53%), Positives = 52/67 (77%), Gaps = 0/67 (0%)
Query 106 DRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENESIR 165
DR+ IHSQL+HLQSKY GTG+ADT++ +W +N RD+ S++GH+ L YFA++ENES
Sbjct 3 DRYNIHSQLEHLQSKYIGTGHADTSKWEWLVNQHRDSYCSYMGHFDLLNYFAISENESKA 62
Query 166 RIRYRCL 172
R+R+ +
Sbjct 63 RVRFNLM 69
> cpv:cgd4_890 hypothetical protein ; K12832 splicing factor 3B
subunit 5
Length=85
Score = 73.6 bits (179), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 37/72 (51%), Positives = 50/72 (69%), Gaps = 1/72 (1%)
Query 102 MSNMDRFTIHSQLQHLQSKYPGTGNADTTRL-DWGINIQRDTLASHVGHYSRLAYFAVAE 160
M +R TI+ QL+ L SKY GTG+ +T R +WG NI RDT +S++ H SR++YFA+AE
Sbjct 1 MDGENRSTIYFQLEQLHSKYQGTGHINTKRSSEWGDNILRDTASSNIMHQSRISYFAIAE 60
Query 161 NESIRRIRYRCL 172
N S RI +R L
Sbjct 61 NTSKARISFRML 72
> sce:YNL138W-A YSF3, RCP10; Component of the SF3b subcomplex
of the U2 snRNP, essential protein required for for splicing
and for assembly of SF3b; K12832 splicing factor 3B subunit
5
Length=85
Score = 39.3 bits (90), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 118 QSKYPGTGNADTTRLDWGINIQRDTLASHVGHYSRLAYFAVAENE-SIRRIRYRCL 172
+ KY G G+ TTR W N++ DTL + GH + L Y +++ + SIR R L
Sbjct 15 KQKYIGLGDESTTREQWQRNVRNDTLNTLQGHSASLEYVSLSRGDLSIRDTRIHLL 70
> ath:AT1G72320 APUM23; APUM23 (Arabidopsis Pumilio 23); RNA binding
/ binding; K14790 nucleolar protein 9
Length=753
Score = 33.5 bits (75), Expect = 0.34, Method: Composition-based stats.
Identities = 16/45 (35%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 13 QGPFSARKTTAAAAATTPEQQRRQQQHQQ--YNRCCSDFSSSENS 55
QGP+ RK A+ P+Q + +Q+ +Q YN CS F S++++
Sbjct 613 QGPYLLRKLDIDGYASRPDQWKSRQEAKQSTYNEFCSAFGSNKSN 657
> cpv:cgd8_4000 hypothetical protein
Length=2010
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 9 RTKYQGPFSARKTTAAAAATTPEQQRRQQQHQQ 41
+ K + P +KTT A +T E++ R+Q H++
Sbjct 333 KKKMEAPIPPKKTTVAPKSTEVEKEEREQSHEK 365
> cel:B0495.6 hypothetical protein
Length=87
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 21/29 (72%), Gaps = 1/29 (3%)
Query 106 DRFTIHSQLQHLQSKYPGTG-NADTTRLD 133
+RF + +QL+HLQSKY GT + +R+D
Sbjct 6 ERFHVLAQLEHLQSKYTGTAMRHEPSRMD 34
> hsa:2045 EPHA7, EHK3, HEK11; EPH receptor A7 (EC:2.7.10.1);
K05108 EphA7 [EC:2.7.10.1]
Length=998
Score = 30.4 bits (67), Expect = 2.9, Method: Composition-based stats.
Identities = 23/71 (32%), Positives = 32/71 (45%), Gaps = 13/71 (18%)
Query 34 RRQQQHQQYNRCCS-DFSSSENSTR-----------SCPPCLCLTWRPAIPVNLLFACRK 81
+ Q Q +RC + FS E S+R S PP + T P+ P NL+F +
Sbjct 285 KSSSQDLQCSRCPTHSFSDKEGSSRCECEDGYYRAPSDPPYVACTRPPSAPQNLIFNINQ 344
Query 82 GLLLL-WSPAA 91
+ L WSP A
Sbjct 345 TTVSLEWSPPA 355
> mmu:72656 Ints8, 2810013E07Rik, AV063769, D130008D20Rik; integrator
complex subunit 8; K13145 integrator complex subunit
8
Length=995
Score = 29.3 bits (64), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 0/81 (0%)
Query 83 LLLLWSPAAATVLLLQQAKMSNMDRFTIHSQLQHLQSKYPGTGNADTTRLDWGINIQRDT 142
L +L AA +L+L+ A N D + + L + PGT N +T G+ I+ +
Sbjct 188 LKVLKEQAADCILVLEAALRLNKDLYVHTMRTLDLLAVEPGTVNGETENSTAGLKIRTEE 247
Query 143 LASHVGHYSRLAYFAVAENES 163
+ V + AYF +S
Sbjct 248 MQCQVCYDLGAAYFQQGSTDS 268
Lambda K H
0.322 0.131 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4341553636
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40