bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1863_orf1
Length=137
Score E
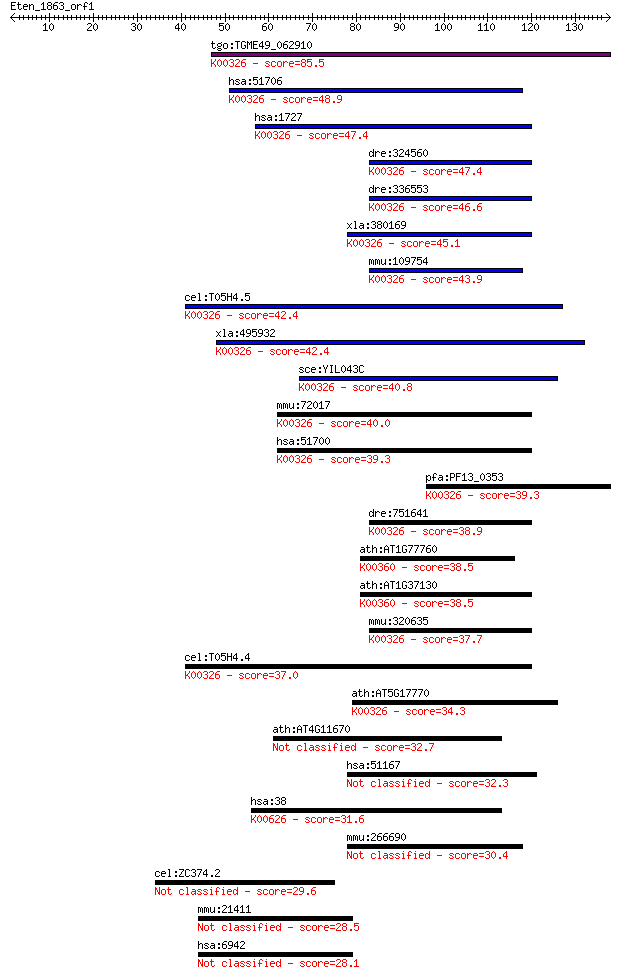
Sequences producing significant alignments: (Bits) Value
tgo:TGME49_062910 NADH-cytochrome B5 reductase, putative (EC:1... 85.5 4e-17
hsa:51706 CYB5R1, B5R.1, B5R1, NQO3A2, humb5R2; cytochrome b5 ... 48.9 5e-06
hsa:1727 CYB5R3, B5R, DIA1; cytochrome b5 reductase 3 (EC:1.6.... 47.4 1e-05
dre:324560 cyb5r3, fc32h03, wu:fc32h03, zgc:77071; cytochrome ... 47.4 2e-05
dre:336553 dia1, CYB5R1, MGC56042, wu:fb13e02, wu:fb15h03, zgc... 46.6 3e-05
xla:380169 cyb5r3, MGC52778, dia1; cytochrome b5 reductase 3 (... 45.1 6e-05
mmu:109754 Cyb5r3, 0610016L08Rik, 2500002N19Rik, C85115, Dia-1... 43.9 2e-04
cel:T05H4.5 hypothetical protein; K00326 cytochrome-b5 reducta... 42.4 4e-04
xla:495932 cyb5r2; cytochrome b5 reductase 2 (EC:1.6.2.2); K00... 42.4 5e-04
sce:YIL043C CBR1, CBR5; Cbr1p (EC:1.6.2.2); K00326 cytochrome-... 40.8 0.001
mmu:72017 Cyb5r1, 1500005G05Rik, B5R.1, C80155, Nqo3a2; cytoch... 40.0 0.002
hsa:51700 CYB5R2, B5R.2; cytochrome b5 reductase 2 (EC:1.6.2.2... 39.3 0.003
pfa:PF13_0353 NADH-cytochrome b5 reductase, putative (EC:1.6.2... 39.3 0.003
dre:751641 cyb5r2, MGC153291, im:7147295, zgc:153291; cytochro... 38.9 0.005
ath:AT1G77760 NIA1; NIA1 (NITRATE REDUCTASE 1); nitrate reduct... 38.5 0.006
ath:AT1G37130 NIA2; NIA2 (NITRATE REDUCTASE 2); nitrate reduct... 38.5 0.007
mmu:320635 Cyb5r2, D630003K02Rik, MGC130177, MGC130178, b5R.2;... 37.7 0.012
cel:T05H4.4 hypothetical protein; K00326 cytochrome-b5 reducta... 37.0 0.020
ath:AT5G17770 ATCBR; ATCBR (ARABIDOPSIS THALIANA NADH:CYTOCHRO... 34.3 0.12
ath:AT4G11670 hypothetical protein 32.7 0.36
hsa:51167 CYB5R4, NCB5OR, RP4-676J13.1, cb5/cb5R, dJ676J13.1; ... 32.3 0.49
hsa:38 ACAT1, ACAT, MAT, T2, THIL; acetyl-CoA acetyltransferas... 31.6 0.83
mmu:266690 Cyb5r4, 2810034J18Rik, B5+B5R, C79736, Cb5cb5r, Ncb... 30.4 1.8
cel:ZC374.2 hypothetical protein 29.6 3.0
mmu:21411 Tcf20, 2810438H08Rik, SPBP, mKIAA0292; transcription... 28.5 6.8
hsa:6942 TCF20, AR1, KIAA0292, SPBP; transcription factor 20 (... 28.1 8.2
> tgo:TGME49_062910 NADH-cytochrome B5 reductase, putative (EC:1.7.1.1);
K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=339
Score = 85.5 bits (210), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 47/100 (47%), Positives = 59/100 (59%), Gaps = 9/100 (9%)
Query 47 VAVAVGVIAVAVAAALRRR---------RAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFV 97
V AV V+ A+ L+ R A PFLDK+RK + LVDK +S +T +F
Sbjct 30 VVAAVTVLVCALHWMLKNRASGKSFFLWSAELATPFLDKSRKKLVLVDKITISPNTFKFR 89
Query 98 FAIDHPQRPLGLPVGQHLKLFAPNAHGQVPGHWNGRPDPE 137
F ++ P + LGLPVG+HLKLFAP G VPGHWN PD E
Sbjct 90 FRLECPSQSLGLPVGKHLKLFAPAPKGTVPGHWNKVPDTE 129
> hsa:51706 CYB5R1, B5R.1, B5R1, NQO3A2, humb5R2; cytochrome b5
reductase 1 (EC:1.6.2.2); K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=305
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 29/68 (42%), Positives = 40/68 (58%), Gaps = 1/68 (1%)
Query 51 VGVIAVAVAAALRRRRAPPLQPFLDKTRKCVT-LVDKSFVSHDTIRFVFAIDHPQRPLGL 109
V ++ +AV + L RR P LD K + L+DK+ VSH+T RF FA+ LGL
Sbjct 18 VTLLGLAVGSYLVRRSRRPQVTLLDPNEKYLLRLLDKTTVSHNTKRFRFALPTAHHTLGL 77
Query 110 PVGQHLKL 117
PVG+H+ L
Sbjct 78 PVGKHIYL 85
> hsa:1727 CYB5R3, B5R, DIA1; cytochrome b5 reductase 3 (EC:1.6.2.2);
K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=301
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/63 (39%), Positives = 36/63 (57%), Gaps = 0/63 (0%)
Query 57 AVAAALRRRRAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLK 116
++ L +R P + + + L+D+ +SHDT RF FA+ PQ LGLPVGQH+
Sbjct 21 SLLMKLFQRSTPAITLESPDIKYPLRLIDREIISHDTRRFRFALPSPQHILGLPVGQHIY 80
Query 117 LFA 119
L A
Sbjct 81 LSA 83
> dre:324560 cyb5r3, fc32h03, wu:fc32h03, zgc:77071; cytochrome
b5 reductase 3 (EC:1.6.2.2); K00326 cytochrome-b5 reductase
[EC:1.6.2.2]
Length=298
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/37 (59%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 83 LVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
LVDK +SHDT RF FA+ P LGLP+GQH+ L A
Sbjct 44 LVDKEIISHDTRRFRFALKSPDHVLGLPIGQHIYLSA 80
> dre:336553 dia1, CYB5R1, MGC56042, wu:fb13e02, wu:fb15h03, zgc:56042;
diaphorase (NADH) (cytochrome b-5 reductase) (EC:1.6.2.2);
K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=304
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 22/37 (59%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 83 LVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
LVDK +SHDT RF FA+ P+ LGLPVG+H+ L A
Sbjct 50 LVDKEIISHDTRRFRFALPSPEHVLGLPVGKHVYLSA 86
> xla:380169 cyb5r3, MGC52778, dia1; cytochrome b5 reductase 3
(EC:1.6.2.2); K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=301
Score = 45.1 bits (105), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 29/42 (69%), Gaps = 0/42 (0%)
Query 78 RKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
+ + L+D+ +SHDT RF FA+ P+ LGLP+GQH+ L A
Sbjct 42 KYALRLIDREEISHDTRRFRFALPSPEHVLGLPIGQHIYLSA 83
> mmu:109754 Cyb5r3, 0610016L08Rik, 2500002N19Rik, C85115, Dia-1,
Dia1, WU:AL591952.1-001, WU:AL591952.1-002, WU:AL591952.1-003,
WU:Cyb5r3; cytochrome b5 reductase 3 (EC:1.6.2.2); K00326
cytochrome-b5 reductase [EC:1.6.2.2]
Length=301
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/35 (57%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 83 LVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKL 117
L+DK +S DT RF FA+ PQ LGLP+GQH+ L
Sbjct 47 LIDKEVISPDTRRFRFALPSPQHILGLPIGQHIYL 81
> cel:T05H4.5 hypothetical protein; K00326 cytochrome-b5 reductase
[EC:1.6.2.2]
Length=309
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 47/98 (47%), Gaps = 13/98 (13%)
Query 41 MLLDWRVAVAVGVIAVAVAAALRRRRAPPLQPF-LDKTRKCVTLVDKSF----------- 88
M+ + + + GV+ V+ A +A PF L K K TLVD S
Sbjct 1 MVENNTLIITGGVVVVSSIAIYIYLKATNTCPFSLCKKSKKRTLVDDSVKYPLPLIEKFE 60
Query 89 VSHDTIRFVFAIDHPQRPLGLPVGQHLKLFAPNAHGQV 126
+SHDT +F F + LGLP+GQH+ L A N G++
Sbjct 61 ISHDTRKFRFGLPSKDHILGLPIGQHVYLTA-NIDGKI 97
> xla:495932 cyb5r2; cytochrome b5 reductase 2 (EC:1.6.2.2); K00326
cytochrome-b5 reductase [EC:1.6.2.2]
Length=296
Score = 42.4 bits (98), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 44/96 (45%), Gaps = 12/96 (12%)
Query 48 AVAVGVIAVAVAAALRRRRAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFVFAIDHPQRPL 107
A+ V V+ + A + P+ + + L++K +SHDT +F F + + L
Sbjct 7 AIGVTVLLFLIKALGSGAKKAPVTLLDPNAKYPLPLIEKQEISHDTKKFRFGLPSAEHVL 66
Query 108 GLPVGQHLKL------------FAPNAHGQVPGHWN 131
GLPVGQH+ L + P + +V GH +
Sbjct 67 GLPVGQHIYLSAKVNGSLVVRAYTPVSSDEVKGHVD 102
> sce:YIL043C CBR1, CBR5; Cbr1p (EC:1.6.2.2); K00326 cytochrome-b5
reductase [EC:1.6.2.2]
Length=284
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 36/62 (58%), Gaps = 4/62 (6%)
Query 67 APPLQPFLDKTR---KCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFAPNAH 123
P +P LD R + LV+K+ ++H+T + F + H LGLP+GQH+ + A N +
Sbjct 26 GPKTKPVLDPKRNDFQSFPLVEKTILTHNTSMYKFGLPHADDVLGLPIGQHIVIKA-NIN 84
Query 124 GQ 125
G+
Sbjct 85 GK 86
> mmu:72017 Cyb5r1, 1500005G05Rik, B5R.1, C80155, Nqo3a2; cytochrome
b5 reductase 1 (EC:1.6.2.2); K00326 cytochrome-b5 reductase
[EC:1.6.2.2]
Length=305
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 34/58 (58%), Gaps = 0/58 (0%)
Query 62 LRRRRAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
+RR R P + + + L+DK+ VSH+T RF FA+ LGLPVG+H+ L A
Sbjct 30 VRRSRRPQVTLQDPDEKYLLRLLDKTTVSHNTRRFRFALPTAHHILGLPVGKHVYLSA 87
> hsa:51700 CYB5R2, B5R.2; cytochrome b5 reductase 2 (EC:1.6.2.2);
K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=276
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 0/58 (0%)
Query 62 LRRRRAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
+ RR P+ + + + L++K +SH+T RF F + P LGLPVG +++L A
Sbjct 1 MNSRRREPITLQDPEAKYPLPLIEKEKISHNTRRFRFGLPSPDHVLGLPVGNYVQLLA 58
> pfa:PF13_0353 NADH-cytochrome b5 reductase, putative (EC:1.6.2.2);
K00326 cytochrome-b5 reductase [EC:1.6.2.2]
Length=362
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 0/42 (0%)
Query 96 FVFAIDHPQRPLGLPVGQHLKLFAPNAHGQVPGHWNGRPDPE 137
F+F+ LGL + +H+K A N G++ G WN D E
Sbjct 97 FIFSYPDEYEYLGLGICKHIKFNASNIEGKIKGKWNNNDDKE 138
> dre:751641 cyb5r2, MGC153291, im:7147295, zgc:153291; cytochrome
b5 reductase 2 (EC:1.6.2.2); K00326 cytochrome-b5 reductase
[EC:1.6.2.2]
Length=309
Score = 38.9 bits (89), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 83 LVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
L++K ++HDT RF F + LGLP+GQH+ L A
Sbjct 55 LIEKEEINHDTKRFRFGLPSSSHVLGLPIGQHIYLSA 91
> ath:AT1G77760 NIA1; NIA1 (NITRATE REDUCTASE 1); nitrate reductase
(EC:1.7.1.1); K00360 nitrate reductase (NADH) [EC:1.7.1.1]
Length=917
Score = 38.5 bits (88), Expect = 0.006, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 81 VTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHL 115
V L++K+ +SHD +F FA+ + LGLPVG+H+
Sbjct 665 VRLIEKTSISHDVRKFRFALPSEDQQLGLPVGKHV 699
> ath:AT1G37130 NIA2; NIA2 (NITRATE REDUCTASE 2); nitrate reductase
(NADH)/ nitrate reductase (EC:1.7.1.1); K00360 nitrate
reductase (NADH) [EC:1.7.1.1]
Length=917
Score = 38.5 bits (88), Expect = 0.007, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 81 VTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
V LV+K+ +SHD +F FA+ LGLPVG+H+ L A
Sbjct 665 VQLVEKTSISHDVRKFRFALPVEDMVLGLPVGKHIFLCA 703
> mmu:320635 Cyb5r2, D630003K02Rik, MGC130177, MGC130178, b5R.2;
cytochrome b5 reductase 2 (EC:1.6.2.2); K00326 cytochrome-b5
reductase [EC:1.6.2.2]
Length=292
Score = 37.7 bits (86), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 17/37 (45%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 83 LVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFA 119
L++K +SH+T RF F + P LGLPVG ++ L A
Sbjct 22 LIEKEQISHNTRRFRFGLPSPDHVLGLPVGNYVHLLA 58
> cel:T05H4.4 hypothetical protein; K00326 cytochrome-b5 reductase
[EC:1.6.2.2]
Length=303
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 46/87 (52%), Gaps = 10/87 (11%)
Query 41 MLLDWRVAVAVGVI---AVAVAAALRRRRAP-----PLQPFLDKTRKCVTLVDKSFVSHD 92
M+ + +A+ GV+ +V++ LR+ RA L+ D + + L++K +SH+
Sbjct 1 MVENNTLAITGGVVLISSVSLFLYLRQLRAEKKSKRTLED--DSVKYLLPLIEKFEISHN 58
Query 93 TIRFVFAIDHPQRPLGLPVGQHLKLFA 119
T +F F + LGLP+G H+ L A
Sbjct 59 TRKFRFGLPSKDHILGLPIGHHVYLSA 85
> ath:AT5G17770 ATCBR; ATCBR (ARABIDOPSIS THALIANA NADH:CYTOCHROME
B5 REDUCTASE 1); cytochrome-b5 reductase; K00326 cytochrome-b5
reductase [EC:1.6.2.2]
Length=281
Score = 34.3 bits (77), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 79 KCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFAPNAHGQ 125
K LV + +SH+ +FVF + LGLP+GQH+ + G+
Sbjct 48 KEFKLVKRHQLSHNVAKFVFELPTSTSVLGLPIGQHISCRGKDGQGE 94
> ath:AT4G11670 hypothetical protein
Length=1117
Score = 32.7 bits (73), Expect = 0.36, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query 61 ALRRRRAPPLQPFLDKTR-----KCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVG 112
+L+R+ P LQP + T K T + ++ V T+R ID PQ LGL VG
Sbjct 212 SLKRKDKPHLQPQISNTHSEISSKMDTCIRRNLVQLATLRTGEQIDLPQLALGLLVG 268
> hsa:51167 CYB5R4, NCB5OR, RP4-676J13.1, cb5/cb5R, dJ676J13.1;
cytochrome b5 reductase 4 (EC:1.6.2.2)
Length=521
Score = 32.3 bits (72), Expect = 0.49, Method: Composition-based stats.
Identities = 18/43 (41%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 78 RKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKLFAP 120
RKC L+ K V+HDT F + P L +P+GQH+ L P
Sbjct 276 RKC-QLISKEDVTHDTRLFCLMLP-PSTHLQVPIGQHVYLKLP 316
> hsa:38 ACAT1, ACAT, MAT, T2, THIL; acetyl-CoA acetyltransferase
1 (EC:2.3.1.9); K00626 acetyl-CoA C-acetyltransferase [EC:2.3.1.9]
Length=427
Score = 31.6 bits (70), Expect = 0.83, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 31/57 (54%), Gaps = 0/57 (0%)
Query 56 VAVAAALRRRRAPPLQPFLDKTRKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVG 112
+AV AAL R A P L + + + V++S+VS T++ V + + P+G +G
Sbjct 1 MAVLAALLRSGARSRSPLLRRLVQEIRYVERSYVSKPTLKEVVIVSATRTPIGSFLG 57
> mmu:266690 Cyb5r4, 2810034J18Rik, B5+B5R, C79736, Cb5cb5r, Ncb5or,
b5/b5r, b5b5r; cytochrome b5 reductase 4 (EC:1.6.2.2)
Length=528
Score = 30.4 bits (67), Expect = 1.8, Method: Composition-based stats.
Identities = 17/40 (42%), Positives = 23/40 (57%), Gaps = 2/40 (5%)
Query 78 RKCVTLVDKSFVSHDTIRFVFAIDHPQRPLGLPVGQHLKL 117
R+C L+ K V+HDT F + P L +PVGQH+ L
Sbjct 283 RRC-QLISKEDVTHDTRLFCLMLP-PSTHLQVPVGQHVYL 320
> cel:ZC374.2 hypothetical protein
Length=684
Score = 29.6 bits (65), Expect = 3.0, Method: Composition-based stats.
Identities = 13/41 (31%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 34 RTARMHEMLLDWRVAVAVGVIAVAVAAALRRRRAPPLQPFL 74
+T R +++ DW + V V +V +A +R PP QP +
Sbjct 92 KTERGAQIVEDWTTEIVVDVAKKSVRSAPKRLAPPPSQPIV 132
> mmu:21411 Tcf20, 2810438H08Rik, SPBP, mKIAA0292; transcription
factor 20
Length=1987
Score = 28.5 bits (62), Expect = 6.8, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 44 DWRVAVAVGVIAVAVAAALRRRRAPPLQPFLDKTR 78
DW + A GVIA A R++P Q FLD+ R
Sbjct 1127 DWASSSAQGVIAAAQHRQEGPRKSPRQQQFLDRVR 1161
> hsa:6942 TCF20, AR1, KIAA0292, SPBP; transcription factor 20
(AR1)
Length=1960
Score = 28.1 bits (61), Expect = 8.2, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 44 DWRVAVAVGVIAVAVAAALRRRRAPPLQPFLDKTR 78
DW A GVIA A R++P Q FLD+ R
Sbjct 1099 DWSSGSAQGVIAAAQHRQEGPRKSPRQQQFLDRVR 1133
Lambda K H
0.325 0.139 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2428006156
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40