bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
164,496 sequences; 82,071,388 total letters
Query= Eten_1857_orf1
Length=287
Score E
Sequences producing significant alignments: (Bits) Value
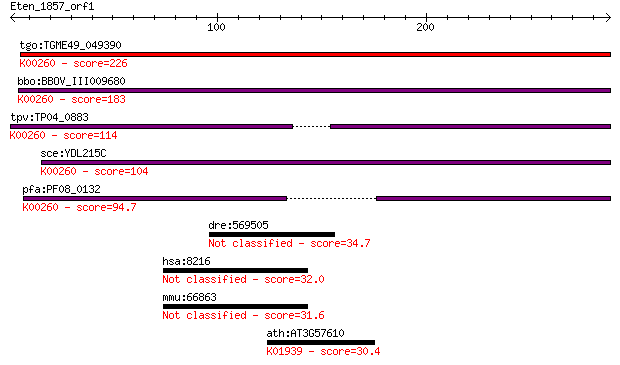
tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putati... 226 7e-59
bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/... 183 6e-46
tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2... 114 3e-25
sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydro... 104 4e-22
pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2); ... 94.7 3e-19
dre:569505 NLR family, pyrin domain containing 1-like 34.7 0.43
hsa:8216 LZTR1, LZTR-1, MGC21205; leucine-zipper-like transcri... 32.0 2.4
mmu:66863 Lztr1, 1200003E21Rik, AI591627, AW550890; leucine-zi... 31.6 3.8
ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylos... 30.4 9.0
> tgo:TGME49_049390 NAD-specific glutamate dehydrogenase, putative
(EC:1.4.1.2); K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1113
Score = 226 bits (576), Expect = 7e-59, Method: Compositional matrix adjust.
Identities = 117/284 (41%), Positives = 175/284 (61%), Gaps = 11/284 (3%)
Query 6 DELPN-PQLSQEQRIDHLLTDLRILFTMPKTKFTNLARERLLTVQETAYALCACQFALHF 64
D+L + P++S +R++ L+ +R+ + M KFT LA+ER LTV E AYA +F +HF
Sbjct 366 DQLTDFPEMSMSRRVELLVKAIRMQYIMQPCKFTELAQERRLTVHEAAYAWAVSKFIVHF 425
Query 65 SATVGSSFPRIEKLVKQFGGKS-LTMQEVYELRTRLKVCPFTEDLIYKVVKENPQMLKLL 123
S TVG +F IEK+VK FG + L+ QE+YELRTRLK+ PF+ED + KVV +P M+K L
Sbjct 426 SGTVGPAFGAIEKMVKHFGSATHLSHQELYELRTRLKLPPFSEDTVLKVVDSHPDMIKRL 485
Query 124 YKEFESLHNPRVARRCAAPSSAAAAAAAAAAAAAASPLRGLVAQLDAFEAVPVFMKFLEF 183
Y+EF+ +H+PR + A + L+ + LD+ +A P+ +KF F
Sbjct 486 YEEFQEMHHPR---------AYAERGHVLKNWEEETSLKRDIQCLDSPDAPPILLKFRLF 536
Query 184 NFSTLKTNFFVPAKCGFGFRLCGEFLPKEDFENKPFSVVFHLAAAALGLHVRCSEVARGG 243
N ++TNF+ K FR+ FLP+ D+ +P++++F + G H R ++VARGG
Sbjct 537 NKHIVRTNFWKDVKQALAFRMDTSFLPEADYPERPYAILFTVGTNFTGFHARFADVARGG 596
Query 244 LRLVRSVGFGVYVQNARALVDEVFRLAFTQHFKNKDLSEGGSKG 287
+R+V+S Y +N DEV++LA TQ+ KNKD+ EGGSKG
Sbjct 597 VRVVQSFTTQSYQRNRDTAFDEVYKLASTQNLKNKDIPEGGSKG 640
> bbo:BBOV_III009680 17.m07840; Glutamate/Leucine/Phenylalanine/Valine
dehydrogenase family protein (EC:1.4.1.2); K00260 glutamate
dehydrogenase [EC:1.4.1.2]
Length=1025
Score = 183 bits (465), Expect = 6e-46, Method: Composition-based stats.
Identities = 101/283 (35%), Positives = 153/283 (54%), Gaps = 15/283 (5%)
Query 5 TDELPNPQLSQEQRIDHLLTDLRILFTMPKTKFTNLARERLLTVQETAYALCACQFALHF 64
+ EL NP LS R ++T +R+ +P + + ++ ERLL E AYA CA F HF
Sbjct 300 STELDNPDLSLLDRAHSIMTAIRLCGILPHSHYLSILTERLLNGSEMAYAYCASIFVEHF 359
Query 65 SATVGSSFPRIEKLVKQFGGKSLTMQEVYELRTRLKVCPFTEDLIYKVVKENPQMLKLLY 124
S +VG IE+L + + + E+Y++R++L + + + I++ V+EN +LK L+
Sbjct 360 SGSVGPHMALIERLATR---EQMAPSELYDIRSKLMIPSYLPNQIFEAVQENIPILKQLH 416
Query 125 KEFESLHNPRVARRCAAPSSAAAAAAAAAAAAAASPLRGLVAQLDAFEAVPVFMKFLEFN 184
K F LHNP + P+ + A L+ + LD + FL FN
Sbjct 417 KNFCILHNPDIN-----PNGTNTDPDSNA-------LKETIKTLDNQVHAKILSLFLTFN 464
Query 185 FSTLKTNFFVPAKCGFGFRLCGEFLPKEDFENKPFSVVFHLAAAALGLHVRCSEVARGGL 244
STL+TNFFV K F FRL FL K D+ P+ +V + G H+R SE++RGG+
Sbjct 465 SSTLRTNFFVTEKSSFAFRLDPSFLSKNDYPETPYGIVMLMGPFFRGFHIRFSEISRGGI 524
Query 245 RLVRSVGFGVYVQNARALVDEVFRLAFTQHFKNKDLSEGGSKG 287
R+V+S + +N + DE + L++TQ KNKD+ EGGSKG
Sbjct 525 RVVQSFSHEAFTRNKLQVFDEAYNLSYTQSLKNKDIPEGGSKG 567
> tpv:TP04_0883 NAD-specific glutamate dehydrogenase (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1178
Score = 114 bits (286), Expect = 3e-25, Method: Composition-based stats.
Identities = 53/134 (39%), Positives = 79/134 (58%), Gaps = 0/134 (0%)
Query 154 AAAAASPLRGLVAQLDAFEAVPVFMKFLEFNFSTLKTNFFVPAKCGFGFRLCGEFLPKED 213
A + A + + QL+ E + + + F++FN TL+TNFFVP K FR FL K D
Sbjct 522 AESLAKKIIRTIKQLENLEEIDILLYFIKFNNHTLRTNFFVPNKISLSFRFNCGFLSKLD 581
Query 214 FENKPFSVVFHLAAAALGLHVRCSEVARGGLRLVRSVGFGVYVQNARALVDEVFRLAFTQ 273
+ P+ + + +G H+R SE++RGG+R+V+S Y +N + DE + L+FTQ
Sbjct 582 YPKVPYGIALIIGPHFMGFHIRFSEISRGGVRVVQSFSEEAYTRNKLQIFDEAYNLSFTQ 641
Query 274 HFKNKDLSEGGSKG 287
KNKD+ EGGSKG
Sbjct 642 SLKNKDIPEGGSKG 655
Score = 86.3 bits (212), Expect = 1e-16, Method: Composition-based stats.
Identities = 41/135 (30%), Positives = 77/135 (57%), Gaps = 8/135 (5%)
Query 1 PAASTDELPNPQLSQEQRIDHLLTDLRILFTMPKTKFTNLARERLLTVQETAYALCACQF 60
P S +LP ++R+ L+ + + +P +K + L +R+L+ E+ Y+ C F
Sbjct 299 PNQSKIDLP-----IKERMSSLIDSIVLSSIVPSSKISILNVDRVLSFYESTYSYCVSTF 353
Query 61 ALHFSATVGSSFPRIEKLVKQFGGKSLTMQEVYELRTRLKVCPFTEDLIYKVVKENPQML 120
HFS +VG ++ + K ++ E++E++++LK+ P+T IY + NP+++
Sbjct 354 IQHFSGSVGPYVSTVDNMAKN---NQVSQSELHEIKSKLKIQPYTPLQIYTAISNNPEIV 410
Query 121 KLLYKEFESLHNPRV 135
K+LYK FE LHNP++
Sbjct 411 KMLYKHFEILHNPKL 425
> sce:YDL215C GDH2; Gdh2p (EC:1.4.1.2); K00260 glutamate dehydrogenase
[EC:1.4.1.2]
Length=1092
Score = 104 bits (259), Expect = 4e-22, Method: Composition-based stats.
Identities = 79/286 (27%), Positives = 134/286 (46%), Gaps = 25/286 (8%)
Query 16 EQRIDHLLTDLRILFTMPKTKFTNLARERLLTVQETAYALCACQFALHFSATVGSSFPRI 75
E + + + +L+ +P F + + R + +E YA F HF +GS +
Sbjct 353 EAALKQVEREASLLYAIPNNSFHEVYQRRQFSPKEAIYAHIGAIFINHFVNRLGSDY--- 409
Query 76 EKLVKQFGGK--SLTMQEVYE-LRTRLKVCPFTEDLIYKVVKENPQMLKLLYKEFESLHN 132
+ L+ Q K T+ E+ E L+ +L+ T+ I ++ ++ ++ LYK F +H
Sbjct 410 QNLLSQITIKRNDTTLLEIVENLKRKLRNETLTQQTIINIMSKHYTIISKLYKNFAQIHY 469
Query 133 PRVARRCAAPSSAAAAAAAAAAAAAASPLRG---LVAQLDAFEAVP------VFMKFLE- 182
+ + + P + A L+ F +P + +K L
Sbjct 470 YHNS------TKDMEKTLSFQRLEKVEPFKNDQEFEAYLNKF--IPNDSPDLLILKTLNI 521
Query 183 FNFSTLKTNFFVPAKCGFGFRLCGEF-LPKEDFENKPFSVVFHLAAAALGLHVRCSEVAR 241
FN S LKTNFF+ K FRL + K ++ P+ + F + G H+R ++AR
Sbjct 522 FNKSILKTNFFITRKVAISFRLDPSLVMTKFEYPETPYGIFFVVGNTFKGFHIRFRDIAR 581
Query 242 GGLRLVRSVGFGVYVQNARALVDEVFRLAFTQHFKNKDLSEGGSKG 287
GG+R+V S +Y N++ ++DE ++LA TQ KNKD+ EGGSKG
Sbjct 582 GGIRIVCSRNQDIYDLNSKNVIDENYQLASTQQRKNKDIPEGGSKG 627
> pfa:PF08_0132 glutamate dehydrogenase, putative (EC:1.4.1.2);
K00260 glutamate dehydrogenase [EC:1.4.1.2]
Length=1397
Score = 94.7 bits (234), Expect = 3e-19, Method: Composition-based stats.
Identities = 45/112 (40%), Positives = 65/112 (58%), Gaps = 0/112 (0%)
Query 176 VFMKFLEFNFSTLKTNFFVPAKCGFGFRLCGEFLPKEDFENKPFSVVFHLAAAALGLHVR 235
+ F F LKTNFF+ K G L +E +P+S++ L +G H+R
Sbjct 529 ILQYFYMFEKYALKTNFFLTHKISLAVAFDGALLKDSIYEAQPYSIIMILGLHFVGFHIR 588
Query 236 CSEVARGGLRLVRSVGFGVYVQNARALVDEVFRLAFTQHFKNKDLSEGGSKG 287
S+++RGG+R+V S Y+ N+ L DE + LA+TQ+FKNKD+ EGGSKG
Sbjct 589 FSKISRGGVRIVISNNVNSYMHNSDNLFDEAYNLAYTQNFKNKDIPEGGSKG 640
Score = 49.3 bits (116), Expect = 2e-05, Method: Composition-based stats.
Identities = 32/140 (22%), Positives = 63/140 (45%), Gaps = 14/140 (10%)
Query 7 ELPNPQLSQEQRIDHLLTDLRILFTMPKTKFTNLARERLLTVQETAYALCACQFALHFSA 66
E +L + +I ++ L+ L +KF L+ +R T QE+AY +F FS
Sbjct 302 EREKQKLDLKDKIYKVVKSLKTLCLFNDSKFIQLSVKRTFTAQESAYLFMIIKFITFFST 361
Query 67 TVGSSFPRIEKL--VKQFGGKSL------------TMQEVYELRTRLKVCPFTEDLIYKV 112
SS+ +E ++ + + + +VY ++ +LK +T++ I +
Sbjct 362 NTLSSYKNVEHALNLRNYNNNIMDTTTNSSSSPSSVLNDVYIIKEKLKSSKYTKEEILRC 421
Query 113 VKENPQMLKLLYKEFESLHN 132
+ N + +K+L+ FE N
Sbjct 422 AQSNVRTIKMLFANFEKKLN 441
> dre:569505 NLR family, pyrin domain containing 1-like
Length=919
Score = 34.7 bits (78), Expect = 0.43, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 31/63 (49%), Gaps = 3/63 (4%)
Query 96 RTRLKVCPFTE---DLIYKVVKENPQMLKLLYKEFESLHNPRVARRCAAPSSAAAAAAAA 152
+ RL+VC TE D + KV+ NP LK+L + ++ +P V CAA A
Sbjct 752 KARLRVCGITEAGSDHLGKVLSLNPSHLKVLDLSWNTIGDPGVKHLCAALEKVDCALETL 811
Query 153 AAA 155
A A
Sbjct 812 ALA 814
> hsa:8216 LZTR1, LZTR-1, MGC21205; leucine-zipper-like transcription
regulator 1
Length=840
Score = 32.0 bits (71), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Query 74 RIEKLVKQFGGKSLTMQEVY---ELRTRLKVCPFTEDLIYKVVKENPQMLKLLYKEFESL 130
R+E+L +Q+ S+ +Q V E RL++ E + VVKE+ ++ KEFE L
Sbjct 561 RLEQLCRQYIEASVDLQNVLVVCESAARLQLSQLKEHCLNFVVKESHFNQVIMMKEFERL 620
Query 131 HNP---RVARRCAAP 142
+P + RR P
Sbjct 621 SSPLIVEIVRRKQQP 635
> mmu:66863 Lztr1, 1200003E21Rik, AI591627, AW550890; leucine-zipper-like
transcriptional regulator, 1
Length=837
Score = 31.6 bits (70), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 23/75 (30%), Positives = 38/75 (50%), Gaps = 6/75 (8%)
Query 74 RIEKLVKQFGGKSLTMQEVY---ELRTRLKVCPFTEDLIYKVVKENPQMLKLLYKEFESL 130
R+E+L +Q+ S+ +Q V E RL++ E + +VKE+ ++ KEFE L
Sbjct 558 RLEQLCRQYIEASVDLQNVLVVCESAARLQLGQLKEHCLNFIVKESHFNQVIMMKEFERL 617
Query 131 HNP---RVARRCAAP 142
+P + RR P
Sbjct 618 SSPLIVEIVRRKQQP 632
> ath:AT3G57610 ADSS; ADSS (ADENYLOSUCCINATE SYNTHASE); adenylosuccinate
synthase (EC:6.3.4.4); K01939 adenylosuccinate synthase
[EC:6.3.4.4]
Length=490
Score = 30.4 bits (67), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 124 YKEFESLHNPRVARRCAAPSSAAAAAAAAAAAAAASPLRGLVAQLDAFEAV 174
++ + LH+PR C+A A +A+ + AA +AA+ G + L V
Sbjct 21 HRRYPPLHHPRSFVSCSAKRPAVSASLSVAADSAATESLGRIGSLSQVSGV 71
Lambda K H
0.324 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 11230970432
Database: egene_temp_file_orthology_annotation_similarity_blast_database_866
Posted date: Sep 17, 2011 2:57 PM
Number of letters in database: 82,071,388
Number of sequences in database: 164,496
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40